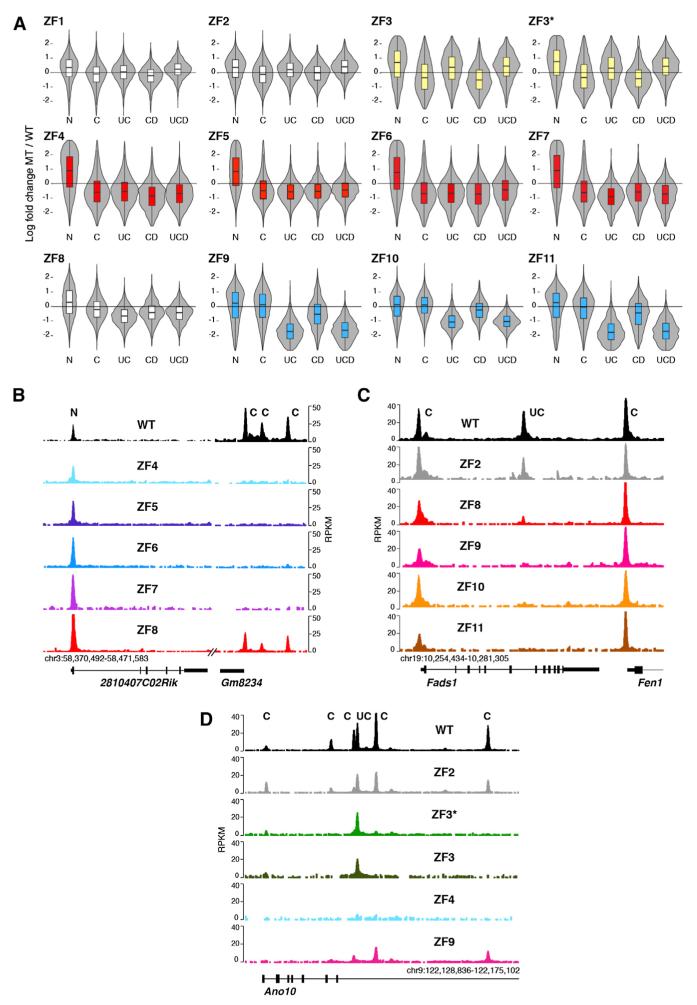
Figure 6. CTCF Uses Different ZF Clusters to Recognize U and C DNA Motifs.
(A) Violin plots showing effects of ZF mutations on CTCF binding sites based on N, C, UC, and UCD classifications described in Figure 4. Data are graphed as the log fold change of mutant to WT ratio. Data were adjusted for global decreases in CTCF binding. Three distinct clusters were highlighted either in yellow (ZF3/3*), red (ZF4–7), or blue (ZF9–11).
(B) Gm8234 mouse locus showing lack of core ZF mutant occupancy at C sites but normal binding to N sites.
(C) Fads1/Fen1 mouse locus depicts defective binding of ZF9–11 mutants to U-containing sites while displaying WT recruitment to sites lacking the motif. ChIP-seq values are plotted as RPKM.
(D) Ano10 locus showing lack of ZF3/3* recruitment to C sites but normal occupancy at binding sites associated with the upstream (U) motif. See also Figure S6.