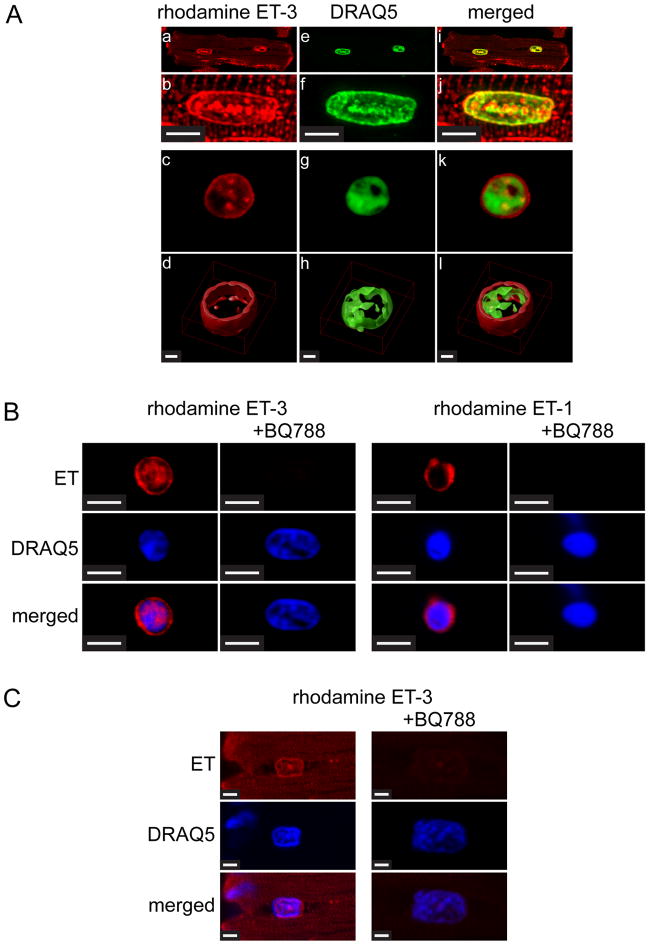
Figure 1. Binding of rhodamine-ET-3 and rhodamine-ET-1 in Triton-skinned ventricular myocytes and nuclei isolated from ventricular myocytes.
A). Freshly isolated adult rat ventricular myocytes were permeabilized with Triton and then plated on laminin-coated Fluorodish culture dishes. Skinned cells were then incubated 30 min with 0.1 μM rhodamine-ET-3 at room temperature. Cells were then washed and DRAQ5, a fluorescent DNA dye was added to stain nuclei. Panels b, f, and j (Scale Bar = 5 μm) show a single nucleus from Panels a, e, and i at higher magnification. Panels c, g and k show an isolated nucleus similarly stained with rhodamine ET-3 and DRAQ5. In panels d, h, and l (Scale Bar = 1 μm) the image stacks for the nuclei shown in the above panels have been deconvolved (Huygens Professional Imaging Software) and then rendered to create 3D images (Volocity 3D Image Analysis Software). B). Nuclei were prepared from freshly isolated adult rat cardiac myocytes, plated on laminin-coated Fluorodish culture dishes and then preincubated with 1 μM ETB-selective antagonist BQ788 for 30 min at room temperature. Rhodamine ET-3 or rhodamine ET-1 (7.5 nM) was added for additional 30 min incubation. Nuclei were then washed 3 times and stained with fluorescent DNA dye, DRAQ5. Scale Bar = 4.8 μm. C). Freshly isolated adult rat ventricular myocytes were permeabilized with Triton and then plated on laminin-coated Fluorodish culture dishes. Skinned cells were then preincubated with 1 μM ETB-selective antagonist BQ788 for 30 min at room temperature. Rhodamine ET-3 (7.5 nM) was added for additional 30 min incubation. Cells were then washed three times and DRAQ5 was added to stain nuclei. Images were acquired using a Zeiss LSM 710 confocal microscope. Scale Bar = 2.3 μm.