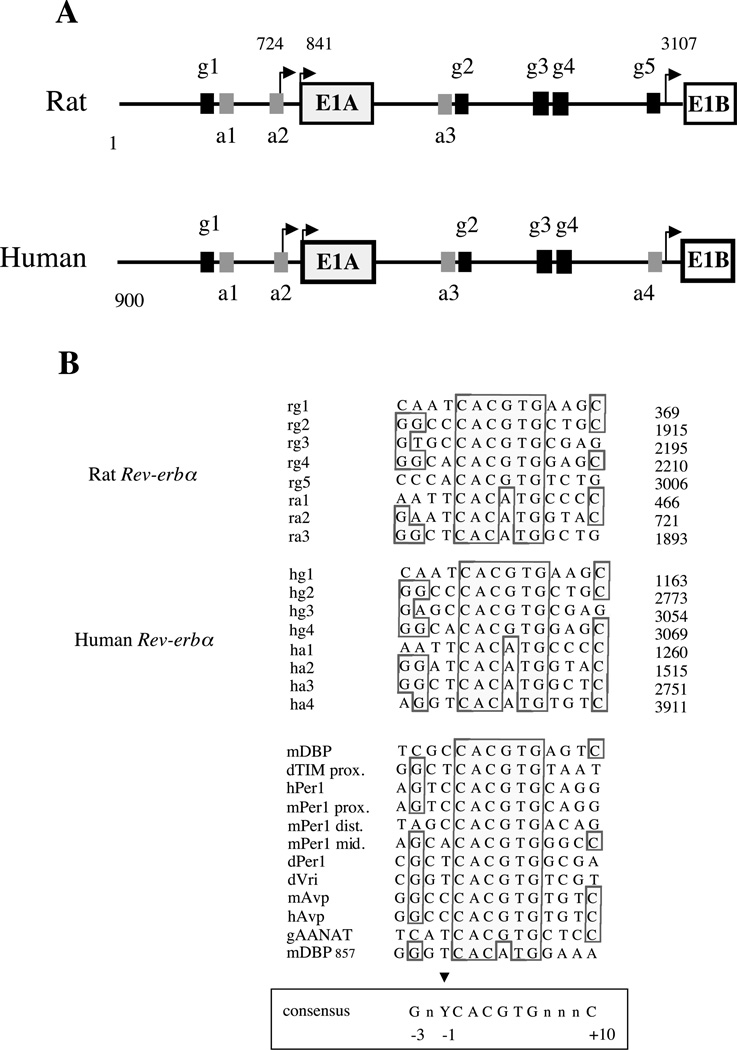
Figure 3.
(A) Genomic structure of Rev-erbα promoters in rat (Rattus norvegicus) and human. P1 and P2 are the two promoters. E1A and E1B are the first alternative exons spliced to exon 2. The numbered E-boxes, g1–g5 (black boxes), correspond to classical E-boxes (CACGTG) and a1–a4 (grey boxes) to the divergent ones (CACATG). Transcription start sites are indicated by arrows. (B) Alignment of the E-boxes found in P1 and P2 rat and human promoters with E-boxes found in various circadian promoters. Numbers on the right indicates the E-boxes position in each promoter. The alignment was obtained by Seqvu software analysis (The Garvan Institute of Medical Research, Sydney, Australia). Boxed bases show conserved positions and the arrow indicates the base similarity at position −1. At −3 and +10 positions were found with a higher frequency of G and C respectively. A consensus sequence derived from this comparison is boxed on the bottom of the Figure.