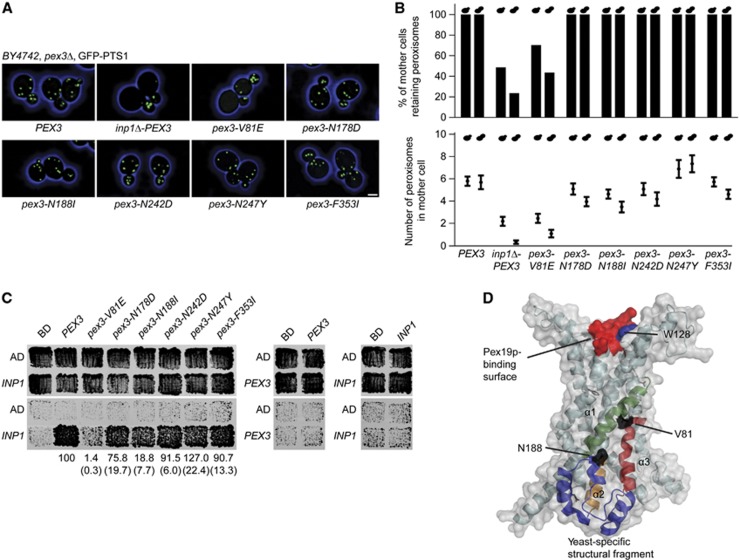
Figure 1.
Cells expressing pex3-V81E have a defect in peroxisome retention. (A) Strain BY4742 lacking the PEX3 gene and expressing the peroxisomal reporter GFP-PTS1 was transformed with plasmids encoding either wild-type PEX3 or mutant pex3 sequences. The strain inp1Δ-PEX3 also carried a deletion of the INP1 gene. Images were acquired by confocal fluorescence microscopy and flattened into maximum intensity projections. Bar, 1 μm. (B) Mother cells were scored for the presence or absence of peroxisomes (upper panel) or total peroxisome numbers (mean±s.e.m., lower panel). Small and large bud size categories are presented in the left and right bars. Quantification was done on at least 100 budded cells of each strain. (C) Yeast two-hybrid analysis to score for interaction between Inp1p and Pex3p. Upper panels show total growth of strains, while bottom panels show growth arising from protein interaction. Strength of interaction between mutant Pex3 proteins and Inp1p in β-galactosidase assays is presented as mean±s.e.m. (brackets) of three independent experiments. (D) Model of Pex3p. Secondary elements likely involved in interacting with Inp1p are highlighted (helices α1, green; α2, orange; and α3, red; yeast-specific structure, blue). The surface of the model is displayed in grey, while the mutated residues V81 and N188 are in black. The approximate binding site for Pex19p is shown in red. Mutagenesis of W128 is described below (Figure 5). See Supplementary Movie 1.