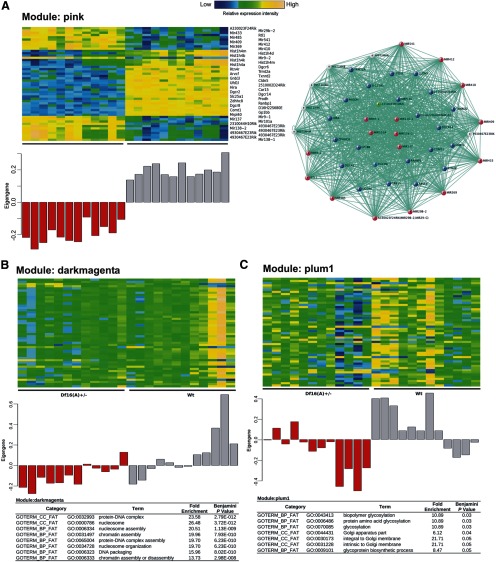
Figure 7.
Disrupted biological processes in the cortex of Df(16)A+/− mice. A, Left, Gene expression heat map of the top 50 genes included in the top module (pink) identified by WGCNA analysis (top) and the module eigengene values (y-axis) for each sample (x-axis). Right, Correlation expression network of the top module. Blue spots indicate genes within the Df16(A) deficiency. Red spots indicate primary transcripts of miRNA genes. Cyan spots represent downregulated protein encoding genes. Yellow spots depict upregulated protein encoding genes. The green spot indicates 2310044H10Rik/Mirta22 (Xu et al., 2013). B, C, Indicated are gene expression heat maps of the top 50 genes included in two modules that are highly correlated with genotype and show statistically significant GO term enrichment (top) and the module eigengene values (y-axis) for each sample (x-axis).