Abstract
Psoriasis is a chronic inflammatory disease of the skin. The causes of psoriasis are unknown, although family and twin studies have shown genetic factors to play a key role in its development. The many genes associated with psoriasis and the immune response include TNFα, IL23, and IL12. Advances in knowledge of the pathogenesis of psoriasis have enabled the development of new drugs that target cytokines (e.g., etanercept, adalimumab, and infliximab, which target TNFα, and ustekinumab, which targets the p40 subunit of IL23 and IL12). These drugs have improved the safety and efficacy of treatment in comparison with previous therapies. However, not all patients respond equally to treatment, possibly owing to interindividual genetic variability. In this review, we describe the genes associated with psoriasis and the immune response, the biological drugs used to treat chronic severe plaque psoriasis, new drugs in phase II and III trials, and current knowledge on the implications of pharmacogenomics in predicting response to these treatments.
1. Introduction
Psoriasis is a chronic inflammatory disease of the skin which is characterized by the presence of erythematous scaly plaques [1]. The prevalence of psoriasis is 2-3% worldwide [2]. Psoriasis has a negative impact on the patient's health and quality of life, is associated with serious medical comorbidities, and affects the quality of life of family members [3, 4].
While the exact cause of psoriasis is unknown, genetic and environmental factors play an important role in its development [5].
The environmental factors that appear to influence the course of and the susceptibility to psoriasis include chronic infections, stress, low humidity, drugs (beta-blockers, lithium, antimalarial agents, and interferon), smoking, and obesity [6].
The role of genetics in the pathogenesis of the disease is well documented in family and twin studies [7]. Genetic factors have been well studied in candidate-driven gene-specific studies and in genomewide association studies (GWAS). The genome regions most strongly associated with the development of the disease are associated with the immune system. Interleukin 23 receptor (IL23R), IL12B, and the human leukocyte antigen Cw6 (HLA-Cw6) of the major histocompatibility complex have been strongly associated with psoriasis [8]. Several studies have described the important role of single-nucleotide polymorphisms (SNPs) in the promoter region of the tumour necrosis factor gene (TNFα) [8].
Discovery of such consistent associations has enabled the development of new, more effective drugs with various targets, such as the p40 subunit of IL-12/23 (ustekinumab) and TNFα (infliximab, adalimumab, and etanercept) [1]. Other biological drugs are in phase III trials and include those targeting IL17 (ixekizumab and secukinumab) and the IL17 receptor (anti-IL17R) (brodalumab), all of which are administered subcutaneously [9]. Phase II clinical trials have demonstrated the efficacy and safety of inhibitors of Janus kinase (JAK) (tofacitinib) and phosphodiesterase 4 (PDE4) (apremilast) [3, 10–13], which are administered orally and may be less expensive than biological drugs.
Although these new drugs have improved tolerability and response to treatment, researchers must increase their knowledge of psoriasis in order to find additional options for oral treatment that are safer, more effective, and free of serious side effects. The influence of genetic polymorphisms on the response to biological drugs has been demonstrated in psoriasis [14, 15]; therefore, advances in pharmacogenetics would enable us to tailor treatment.
In this paper, we describe SNPs in genes associated with psoriasis and those associated with the immune response. We also review current knowledge on biological drugs and the impact of polymorphisms on the response to treatment of psoriasis.
2. Genetics of Psoriasis
The immune system plays a key role in psoriasis. Macrophage activation triggers an immune response that releases TNFα, IL1β, IL12, and IL23 [8]. Psoriasis has been associated with genes involved in the immune response, namely, TNFα, IL12B, and IL23R [8]. However, there has also been associated with genes not involved in immune pathways, such as the early differentiation keratinization markers involucrin (IVL) and small proline-rich protein (SPRR). These genes are involved in atypical epidermal cellular organization and differentiation [16] and are upregulated in psoriasis [17]. A review of the genes and SNPs associated with psoriasis and the immune system is presented in Table 1.
Table 1.
Single-nucleotide polymorphisms (SNPs) in genes associated with psoriasis.
| Gene | Role in immune system* | SNP | MAF** | Minor allele | Population | References |
|---|---|---|---|---|---|---|
| IL23R | Encodes a subunit of the receptor required for IL23A signaling. This protein associates constitutively with JAK2 and binds to transcription activator STAT3 | rs7530511 | 0.125 | T | Caucasian, Japanese, Chinese | [33, 34, 36–38, 41] |
| rs2201841 | 0.275 | C | Caucasian | [44, 45] | ||
| rs11209026 | 0.067 | A | Caucasian | [2, 33, 34, 36, 37, 41–43] | ||
| rs11465817 | 0.279 | A | Chinese | [39] | ||
| rs1343152 | 0.357 | C | Chinese | [39] | ||
| rs2066808 | 0.092 | C | Caucasian | [44] | ||
|
| ||||||
| IL10 | Encodes a cytokine produced by monocytes and lymphocytes that downregulates the expression of Th1 cytokines and blocks NF-κB activity. It enhances B-cell survival, proliferation, and antibody production and regulates the JAK-STAT signaling pathway | rs1800896 | 0.467 | A | Caucasian, Egyptian | [25, 60] |
|
| ||||||
| TNFα | Encodes a proinflammatory cytokine produced by macrophages. TNFα is implicated in multiple roles such as cell proliferation, differentiation, and apoptosis | rs1800629 | 0.217 | A | Caucasian, Egyptian, Korean | [19–22, 25–29] |
| rs361525 | 0.131 | A | Caucasian | [19, 20, 22–24, 26–29] | ||
| rs1799724 | 0.158 | A | Caucasian | [14]# | ||
|
| ||||||
| IL12B | IL12B is a cytokine expressed by activated macrophages that serves as an essential inducer of Th1 cell development | rs6887695 | 0.217 | T | Caucasian, Chinese | [33, 36, 37, 39, 41, 42] |
| rs3212227 | 0.225 | C | Caucasian, Japanese, Chinese | [33, 36–38, 40–43, 46, 103] | ||
| rs2082412 | 0.225 | A | Caucasian | [44] | ||
| rs2546890 | 0.438 | G | Caucasian | [45] | ||
|
| ||||||
| GBP6 | Interferon induces GBP that hydrolyzes GTP to both GDP and GMP | rs928655 | 0.288 | G | Caucasian | [42] |
|
| ||||||
| IL6 | Encodes a cytokine that induces inflammatory responses through IL6Rα and maturation of B cells | rs1800795 | 0.467 | G | Egyptian | [25] |
|
| ||||||
| IL13 | Encodes a cytokine produced by activated Th2 that is involved in maturation and differentiation of B cells. IL13 downregulates macrophage activity and inhibits the production of proinflammatory cytokines and chemokines | rs20541 | 0.233 | T | Caucasian | [2, 44, 61] |
| rs848 | 0.242 | T | Caucasian | [61] | ||
| rs1800925 | 0.196 | T | Caucasian | [61] | ||
|
| ||||||
| TNFAIP3 | TNF induces the expression of TNFAIP3, which inhibits NF-κB activation and TNF-mediated apoptosis. TNFAIP3 is involved in cytokine-mediated immune and inflammatory responses | rs610604 | 0.408 | C | Caucasian | [2, 15, 44]# |
| rs6920220 | 0.175 | A | Caucasian | [33, 44, 47] | ||
| rs10499194 | 0.175 | T | Caucasian | [33, 44, 47] | ||
| rs5029939 | 0.042 | G | Caucasian | [44, 47, 48] | ||
| rs2230926 | 0.027 | G | Caucasian | [15]# | ||
|
| ||||||
| TNIP1 | Encodes TNFAIP3 interacting protein 1, which plays a role in the regulation of NF-κB activation | rs17728338 | 0.075 | A | Caucasian | [2, 44] |
|
| ||||||
| IL1RN | IL1RN inhibits IL1 and modulates immune and inflammatory responses | rs397211 | 0.164 | G | Caucasian | [44] |
|
| ||||||
| HLA-C | HLA class I molecules play a central role in the immune system by presenting peptides derived from endoplasmic reticulum lumen | rs12191877 | 0.125 | T | Caucasian | [44, 45, 51] |
| rs10484554 | 0.135 | T | Caucasian, Chinese | [2, 42, 104] | ||
| rs1265181 | 0.258 | C | Chinese | [35, 104] | ||
| rs3134792 | 0.111 | G | Caucasian | [105] | ||
|
| ||||||
| NF-κBIA | Encodes a member of the NF-κB inhibitor family, which interacts with REL dimers to inhibit NF-κB/REL complexes, which are involved in inflammatory responses | rs2145623 | 0.290 | C | Caucasian | [45] |
| rs8016947 | 0.465 | T | Caucasian | [50] | ||
|
| ||||||
| APOE | APOE plays a role in the proliferation of T lymphocytes and protects against some infections in patients with psoriasis [73] | rs429358 | 0.078 | APOE*4 | Caucasian | [75] |
| rs7412 | — | — | Caucasian | [75] | ||
|
| ||||||
| VDR | Encodes the nuclear hormone receptor for vitamin D3, which regulates immune response pathways | rs4516035 | 0.381 | C | Caucasian | [76] |
|
| ||||||
| IFNγ | Encodes a soluble cytokine with antiviral, immunoregulatory, and antitumor properties, and it is a potent activator of macrophages | rs2430561 | — | — | Caucasian | [54] |
|
| ||||||
| IL2 | Encodes a cytokine that is important for the proliferation of T and B lymphocytes | rs2069762 | — | — | Korean | [53] |
|
| ||||||
| IL4 | IL4 is a pleiotropic cytokine involved in the modulation of Th2 immune responses. IL4 receptor also binds to IL13, which may contribute to many overlapping functions of this cytokine and IL13 | rs2243250 | 0.137 | T | Korean | [53] |
|
| ||||||
| IL15 | Encodes a cytokine that regulates T-cell and natural killer activation and proliferation. IL15 also induces the activation of JAK kinases, as well as the phosphorylation and activation of STAT3, STAT5, and STAT6 | rs2857261 | 0.431 | G | Chinese | [69] |
| rs10519613 | 0.102 | A | Chinese | [69] | ||
| rs1057972 | — | — | Chinese | [69] | ||
|
| ||||||
| TNFRSF1B | TNFRSF1B is a TNFα receptor that mediates the recruitment of antiapoptotic proteins | rs1061622 | 0.239 | G | Caucasian, Japanese | [14]# |
|
| ||||||
| MCP1 | MCP1 encodes a cytokine characterized by two cysteines separated by a single amino acid that displays chemotactic activity for monocytes and basophils | rs1024611 | 0.305 | G | Caucasian | [71] |
|
| ||||||
| CTLA4 | Encodes a protein which inhibits T cells | rs3087243 | 0.460 | A | Caucasian | [81]## |
| rs231775 | 0.389 | G | Caucasian | [81]## | ||
|
| ||||||
| DEFB4 | DEFB4 is a member of a family of microbicidal and cytotoxic peptides made by neutrophils | rs2740091 | — | — | Caucasian | [56] |
| rs2737532 | — | — | Caucasian | [56] | ||
|
| ||||||
| STAT4 | In response to cytokines, the STAT proteins are phosphorylated and translocate to the cell nucleus, where they act as transcription activators. STAT transduces IL12, IL23, and IFN type I signals in T lymphocytes and regulates the differentiation of Th cells | rs7574865 | 0.230 | T | Caucasian | [72] |
|
| ||||||
| IL18 | IL18 stimulates production of IFNγ in Th1 | rs187238 | — | — | Japanese | [58] |
|
| ||||||
| IL19 | IL19 is a member of the IL10 cytokine subfamily with a role in inflammatory responses | rs2243188 | 0.230 | A | Caucasian | [64, 68] |
| rs2243158 | 0.085 | C | Caucasian | [64] | ||
|
| ||||||
| IL20 | Encodes a cytokine structurally related to IL10 and transduces its signal through STAT3 in keratinocytes | rs1713239 | 0.177 | G | Chinese | [65] |
| rs2981572 | — | — | Caucasian | [64, 66, 68] | ||
|
| ||||||
| IL20RA | Encodes a receptor for IL20, a cytokine that may be involved in epidermal function | rs1342642 | 0.314 | A | Caucasian | [67] |
| rs1184860 | — | — | Caucasian | [67] | ||
| rs1167846 | 0.246 | T | Caucasian | [67] | ||
| rs1167849 | 0.285 | A | Caucasian | [67] | ||
|
| ||||||
| ERAP1 | Encodes an aminopeptidase involved in trimming HLA class I-binding precursors so that they can be presented on HLA class I | rs151823 | 0.093 | A | Chinese | [52] |
| rs27524 | 0.332 | A | Caucasian | [50] | ||
|
| ||||||
| IL1B | Encodes a cytokine produced by activated macrophages which plays an important role in the inflammatory response | rs16944 | 0.358 | A | Caucasian | [24] |
|
| ||||||
| TRAF3IP2 | Encodes a protein that interacts with TRAF proteins and plays a central role in innate immunity in response to pathogens, inflammatory signals, and stress | rs13210247 | 0.080 | G | Caucasian | [45, 49] |
| rs33980500 | — | — | Caucasian | [45, 49] | ||
| rs13196377 | 0.053 | A | Caucasian | [49] | ||
| rs13190932 | 0.058 | A | Caucasian | [49] | ||
| rs240993 | 0.250 | T | Caucasian | [50] | ||
|
| ||||||
| IL28RA | Encodes a receptor complex that interacts with IL28A, IL28B, and IL29. The expression of these cytokines can be induced by viral infection | rs4649203 | 0.239 | G | Caucasian | [50] |
|
| ||||||
| TYK2 | Encodes a member of the JAK protein family that promulgate cytokine signals by phosphorylating receptor subunits. TYK2 is a component of IFN I and II signaling pathways and may play a role in antiviral immunity | rs12720356 | 0.124 | C | Caucasian | [50] |
|
| ||||||
| IFIH1 | Encodes a protein that mediates induction of IFN response to viral RNA [83] | rs17716942 | 0.195 | C | Caucasian | [50] |
|
| ||||||
| LCE | Encodes a protein that plays a role in skin barrier function [83] | rs4085613 | 0.403 | T | Caucasian | [50] |
| rs4845454 | 0.403 | C | Caucasian | [50] | ||
| rs1886734 | 0.407 | A | Caucasian | [50] | ||
| rs4112788 | 0.403 | A | Caucasian | [50] | ||
| rs6701216 | 0.137 | T | Caucasian | [42] | ||
| rs4112788 | 0.417 | T | Chinese | [85] | ||
|
| ||||||
| ZNF313 | Encodes a protein that is involved in T-cell activation [83] | rs2235617 | 0.432 | G | Caucasian | [50] |
| rs495337 | 0.430 | A | Caucasian | [105] | ||
*Data from NCBI web page [57]; **MAF: minor allele frequency for Caucasian population (data from HapMap web page [106] and Alfred [107]). IL: interleukin; R: receptor; JAK: Janus kinase; STAT: signal transducer and activator of transcription; Th1: type 1 helper T lymphocyte; TNF: tumor necrosis factor; GBP: guanylate-binding protein; GTP: guanosine triphosphate; GDP: guanosine diphosphate; GMP: guanosine monophosphate; TNFAIP: TNF-alpha interacting protein; TNIP1: TNFAIP3 interacting protein; IL1RN: interleukin 1 receptor antagonist; HLA: human leukocyte antigen; NF-κBIA: nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha; REL: v-rel reticuloendotheliosis viral oncogene; APOE: apolipoprotein E; VDR: vitamin D receptor; TNFRSF1: tumor necrosis factor receptor superfamily; MCP: monocyte chemoattractant protein; CTLA4: cytotoxic T lymphocyte-associated protein 4; DEFB4: defensin beta 4A; IFN: interferon; ERAP: endoplasmic reticulum aminopeptidase; TRAF3IP: TRAF3 (TNF receptor-associated factor 3) interacting protein; IRAK: interleukin-1 receptor-associated kinase; TYK: tyrosine kinase; IFIH1: interferon induced with helicase C domain 1; LCE: late cornified envelope; RNF114: ring finger protein 114; #association between psoriasis and response to anti-TNF treatment; ##haplotype GG of rs3087243-rs231775 associated with psoriasis.
T helper 17 (Th17) lymphocytes release IL22 and IL17 (Figure 1), which are highly expressed in psoriatic skin [18]. These lymphocytes also produce IL2, IFNγ, and TNFα (Figure 1) [3]. The proinflammatory cytokine TNFα plays a key role in the pathogenesis of psoriasis [19, 20]. Polymorphisms in the TNFα gene may alter the release of this cytokine in healthy subjects [21]. A study performed in Caucasian patients with early-onset psoriasis showed a strong association with TNFα polymorphisms (rs1800629 and rs361525) (Table 1) [19]. In this sense, a meta-analysis of 18 published case-control studies showed that when the GA + AA genotype was compared with the GG genotype, the risk of psoriasis increased for rs361525 and decreased for rs1800629 in TNFα gene (Table 1) [22]. Kaluza et al. (2000) observed a decrease in TNFα production in peripheral blood mononuclear cells (47 cases and 43 controls) stimulated with mitogens in psoriatic patients who were A allele carriers of rs361525 (TNFα gene) compared to controls [23]. Moreover, the authors found an association between the A allele in rs361525 in the TNFα gene and increased production of TNFα and early onset of psoriasis (Table 1) [24]. A study performed in an Egyptian population (46 cases and 96 controls) revealed an association between SNPs in TNFα (GG allele in rs1800629) and psoriasis (P < 0.05) (Table 1) [25]. However, no significant differences were found in rs1800629 and rs361525 in this gene in Korean patients with psoriasis (n = 103) and controls (n = 125) [26].
Figure 1.
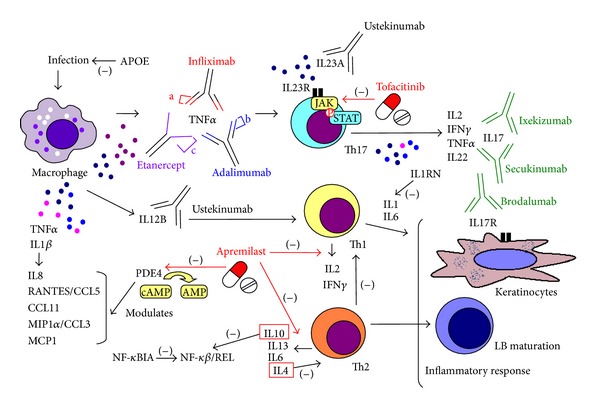
Simplified representation of the main mediators of inflammation in psoriasis, the therapeutic targets of biological drugs, and oral alternatives currently under development. Th: helper T lymphocyte; LB: lymphocyte B; APOE: apolipoprotein E; TNF: tumor necrosis factor; IL: interleukin; RANTES, chemokine regulated on activation normal T cells expressed and secreted; CCL: chemokine Cys-Cys motif ligand; MIP: macrophage inflammatory protein; MCP: monocyte chemoattractant protein; PDE4: phosphodiesterase 4; cAMP: cyclic adenosine monophosphate; IFN: interferon; JAK: Janus kinase; STAT: signal transducer and activator of transcription; NF-κBIA: nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor; NF-κB/REL: nuclear factor kappa B/v-rel reticuloendotheliosis viral oncogene complex; a: infliximab, mouse variable region; b: adalimumab, human variable region; c: etanercept, Human TNRFp75 (TNRF2); (−) indicates inhibition and (→) indicates stimulation.
Reich et al. (1999) analyzed rs361525 and rs1800629 in TNFα gene in patients with type I psoriasis (onset before 40 years; n = 100) and type II psoriasis (onset beyond 40 years; n = 51) and in healthy controls (n = 123) (Table 1) [27]. The results showed that the rs361525*A allele was more frequent and the rs1800629*A allele was less frequent in patients with type I psoriasis than in controls (P = 0.0012 and P = 0.041, resp.), although no differences were found between these polymorphisms and type II psoriasis [27]. Nedoszytko et al. (2007) analyzed 166 patients with psoriasis (134 with type I and 32 with type II) and 65 healthy controls [28] and found similar results to those of Reich et al. [27], with a higher prevalence of the A allele in rs361525 and lower frequency of the A allele in rs1800629 (TNFα gene) in Caucasian patients than in controls (Table 1) [28]. A previous study performed in 99 Caucasian patients (64 with type I psoriasis and 35 with type II psoriasis) showed decreased frequency of the GG genotype and increased frequency of the GA genotype of rs361525 (TNFα gene) in patients with type I psoriasis compared with controls (n = 123) (Table 1) [29]. Therefore, the GG genotype in this SNP is associated with a lower risk of type I disease [29].
The inflammatory response in psoriasis is characterized by production of TNFα, as seen above, and production of IL1β (Figure 1) [24]. In fact, this proinflammatory cytokine is overexpressed in psoriatic lesions [30]. An in vitro study in peripheral blood mononuclear cells (231 cases and 345 controls) revealed an association between the CC genotype in rs16944 in the IL1β gene with increased production of IL1RA in response to lipopolysaccharide and IL10 and late-onset psoriasis (over 40 years) (Table 1) [24]. Johansen et al. (2010) observed that expression of IL1β was decreased 4 days after treatment with adalimumab (a human monoclonal antibody against TNFα) [30].
IL23 regulates and stimulates the activation, differentiation, and survival of Th17 lymphocytes (Figure 1) [31, 32] and is highly expressed in psoriatic lesions [18]. IL12 induces the production of IFNγ by Th1 (Figure 1) [33]. The p40 subunit of IL23 and IL12 is the therapeutic target of ustekinumab, a highly effective biological drug, thus suggesting that IL12 and IL23 play an important role in psoriasis [33–35]. Polymorphisms in IL23R and IL12B have been associated with susceptibility to psoriasis in both Caucasian [36, 37] and Asian patients [38, 39].
In Caucasians, a GWAS (1446 cases and 1432 controls) showed the combination of rs3212227 and rs6887695 in IL12B as a risk haplotype in psoriasis (Table 1) [37]. The authors also found an association between rs11209026 in the IL23R gene and psoriasis [37]. Capon et al. (2007) performed a study of 318 cases and 288 controls and found significant differences between the groups for rs3212227 in IL12B (P = 0.036) (Table 1) [40]. A subsequent GWAS with 1810 cases and 2522 controls found an association between SNPs in IL23R (rs7530511 and rs11209026) and IL12B (rs6887695 and rs3212227) and predisposition to psoriasis in Caucasian patients (Table 1) [36]. Smith et al. (2008) found similar results, associating these four SNPs with psoriasis [41], and Liu et al. (2008) identified an association between psoriasis and IL23R (rs11209026) and IL12B (rs6887695) (Table 1) [42]. Hüffmeier et al. (2009) analyzed the same four SNPs in 1114 patients and found a strong association between rs11209026 (IL23R) and rs3212227 (IL12B) and psoriasis (Table 1) [43]. Another recent study also associated rs11209026 in IL23R gene with psoriasis (Table 1) [2]. Other IL12B and IL23R susceptibility loci identified in GWAS in Caucasian patients include rs2201841 and rs2066808 (IL23R) and rs2082412 and rs2546890 (IL12B) (Table 1) [44, 45].
The SNPs rs11209026 in IL23R gene and rs3212227 in IL12B gene have also been studied in Japanese patients (143 cases and 100 controls), and the A allele (rs3212227) was more frequent in patients with psoriasis than in healthy subjects (Table 1) [46]. In a GWAS performed in a Thai cohort (206 cases and 144 controls), a marginally significant association was found between rs7530511 (IL23R gene) and psoriasis (Table 1) [38]; rs3212227 (IL23R) was also associated with the disease [38]. However, the authors did not find an association with rs6887695 in IL12 gene [38]. A GWAS performed in a Chinese population (217 cases and 288 controls) identified other polymorphisms associated with psoriasis in IL23R (A allele rs11465817-A allele rs1343152 haplotype) and IL12B (rs6887695) (Table 1). The SNP in IL12B was replicated with 578 cases and 1422 controls, and the authors found a positive association with psoriasis [39].
Nair et al. (2009) found strong associations between psoriasis and other genes: IL13, which is involved in Th2 lymphocyte modulation (rs20541); TNFα interacting protein 3 (TNFAIP3) (rs610604, rs6920220, rs10499194, and rs5029939 [47, 48]) and TNFAIP3 interacting protein (TNIP1), which regulate the activity of nuclear factor kappa B (NF-κB) [33]; IL1RN, which inhibits the activity of IL1; and HLA-C (rs12191877), which is involved in inflammatory responses [44] (Table 1). In addition, rs610604 (TNFAIP3) and rs17728338 (TNIP1), but not rs2066808 (IL23R) and rs397211 (IL1RN), were associated with psoriasis in a case-control study (Table 1) [2].
Ellinghaus et al. (2010) studied the TNF receptor-associated factor 3 interacting protein gene (TRAF3IP2) and identified an association between 2 SNPs and psoriasis (rs13210247 and rs33980500) (Table 1) [45]. This association was confirmed by Hüffmeier et al. (2010) in 2040 German patients with psoriasis vulgaris [49]. TRAF3IP2 encodes a protein that interacts with NF-κB/REL (v-rel reticuloendotheliosis viral oncogen) complexes and modulates IL17 pathways [45]. In another GWAS, rs240993 (TRAF3IP2 gene) was associated with psoriasis in Caucasian patients (Table 1) [50]. In the GWAS performed by Ellinghaus et al. (2010), also in Caucasian patients, an association was identified between rs12191877 (HLA-C) and rs2145623 (nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor gene, NF-κBIA) and psoriasis (Table 1) [45]. Feng et al. (2009) performed a GWAS (1359 cases and 1400 controls) and showed rs12191877 (HLA-C) to be a high-risk SNP for psoriasis (Table 1) [51]. The SNP rs8016947 in NF-κBIA was associated with psoriasis (GWAS) (Table 1) [50].
Ellinghaus et al. identified new susceptibility loci [45], such as rs4649203 in IL28RA and rs12720356 in the tyrosine kinase 2 gene (TYK2) (Table 1) [50]. These authors also found an interaction between HLA-C and the endoplasmic reticulum aminopeptidase gene (ERAP1) (rs27524) [50]. In a Chinese population, another SNP in ERAP1 (rs151823) was associated with early-onset psoriasis (less than 40 years) (GWAS, 8312 cases and 12919 controls) (Table 1) [52]. In a case-control study performed in patients with psoriasis (n = 1050; controls n = 1497), the SNPs rs8016947 (NF-κBIA), rs4649203 (IL28RA), rs12720356 (TYR2), and rs27524 (ERAP1) were not associated with the disease [2].
Activation of Th1 lymphocytes was associated with the production of cytokines such as IL2 and INFγ [3, 18] (Figure 1). In a Korean population (114 patients and 281 controls), the rs2069762 (G allele) in IL2 conferred a risk of psoriasis, mainly in the late-onset group (Table 1) [53]. As for INFγ, rs2430561 has been associated with susceptibility to psoriasis (78 cases versus 74 controls) (Table 1) [54]. Furthermore, production of IFNγ was increased by DEFB4 (defensin beta 4A), a microbiocidal and cytotoxic peptide [55]. A significant association was found between rs2740091 and rs2737532 in DEFB4 and predisposition to psoriasis in Caucasian patients (498 cases and 577 controls) (Table 1) [56]. IL18 also stimulates IFNγ production [57], and the presence of polymorphisms in the IL18 gene (rs187238) was associated with susceptibility to psoriasis in Japanese patients (Table 1) [58].
Th2 lymphocytes release IL4, IL6, IL10, and IL13 [3] (Figure 1). A study performed in 114 psoriasis patients and 281 controls from Korea showed that rs2069762 (G allele) in IL2 conferred a risk of developing the disease, mainly in late-onset psoriasis (Table 1) [53]. Moreover, the cytokines IL6 and IL10 seem to be important in the development of psoriasis [59]. In an Egyptian population (46 cases and 96 controls), an association was established between psoriasis and SNPs in IL6 (CC genotype in rs1800795) and IL10 (GG genotype in rs1800896) (Table 1) [25]. In addition, Craven et al. (2001) found differences in rs1800896 (IL10) genotype frequencies between patients with late-onset disease (n = 84) and controls (Table 1) [60]. However, results for the associations between rs1800896 in IL10 gene and psoriasis are controversial, since several studies did not find any differences between cases and controls for this SNP [27, 59]. IL13 is involved in the differentiation and maturation of B cells and differentiation and function of Th17 lymphocytes [33]. Julia et al. (2012) found an association between rs20541 in IL13 and psoriasis (Table 1) [2]. Moreover, the CCG haplotype of rs1800925-rs20541-rs848 in IL13 was associated with susceptibility to psoriasis in a study performed in 1446 cases and 1432 controls (Table 1) [61]. In contrast, Duffin et al. (2009) found these associations with psoriatic arthritis, but not with psoriasis [62], and other authors found that rs20541 and rs1800925 in IL13 gene were involved in psoriatic arthritis but not in psoriasis [63].
Other cytokines and chemokines associated with psoriasis include IL19, IL20, IL15, and MCP1 (monocyte chemoattractant protein). Minor alleles of rs2243188 and rs2243158 in IL19 have a protective effect in patients with the disease (Table 1) [64]. In a case-control study (340 cases and 199 controls), the G allele in rs1713239 (IL20) was associated with psoriasis in a Chinese population (Table 1) [65]. Kingo et al. (2004) found an association between G allele carriers of rs2981572 (IL20) and predisposition to psoriasis in Caucasian patients (Table 1) [66]. Polymorphisms in the IL20 receptor (IL20RA) have also been associated with psoriasis (Table 1) [67]. Of note, the haplotype in IL19 and IL20 exhibited a susceptibility factor for the development of psoriasis [68]. IL15 induces the activation of the Janus kinase/signal transducer transcription activation factor (JAK/STAT) pathway and may trigger an immune response in psoriatic lesions [57, 69]. Polymorphisms in IL15 (rs2857261, rs10519613, and rs1057972) have been associated with psoriasis in a Chinese population (Table 1) [69]. However, in a Caucasian population, no clear association was found between rs1057972 and rs10519613 in IL15 gene and psoriasis [70].
MCP1 is a CC-type chemokine that plays a role in the recruitment of monocytes and T lymphocytes in inflammation [71]. Wang et al. (2008) found high serum levels of MCP1 in patients with psoriasis compared with controls [71]. The SNP rs10224611 (GG or AG genotype) in the MCP1 gene may confer susceptibility to psoriasis (507 cases and 530 controls) (Table 1) [71].
Other genes associated with psoriasis include signal transducer and activator of transcription 4 (STAT4), apoliprotein E (APOE), vitamin D receptor (VDR), and cytotoxic T lymphocyte-associated protein 4 (CTLA4). Zervou et al. (2009) found a weak association between the T allele in rs7574865 (STAT4) and predisposition to psoriasis (Table 1) [72]. APOE may play a role in psoriasis by modifying the proliferation of mitogen-activated T lymphocytes and ensuring protection against some infections (Figure 1) [73]. Other authors have reported the APOE-ε4 allele to be a risk factor for the development of severe form of psoriasis [74]. In addition, 2 SNPs in the APOE gene (rs429358 and rs7412) have been associated with chronic plaque psoriasis and guttate psoriasis (Table 1) [75].
Several authors have demonstrated the role of VDR in the pathogenesis of psoriasis [76, 77]. Rucevic et al. (2009) described possible effects of VDR polymorphisms on the immune system, namely, immunomodulation, stimulation of cellular differentiation, and inhibition of proliferation [78]. The TaqI polymorphism (allele T) in VDR was associated with familial psoriasis in a Turkish population [79]. In addition, the A allele in rs451635 (VDR gene) was protective against susceptibility to nonfamilial psoriasis (Table 1) [76]. In contrast, Zuel-Fakkar et al. (2011) did not find any association between the polymorphisms ApaI and TaqI in VDR and psoriasis [77].
CTLA4 is a protein that downregulates activation of T lymphocytes [80]. The GG haplotype of rs3087243-rs231775 in CTLA4 has been associated with psoriasis, but the analysis of these SNPs individually revealed no statistically significant associations (Table 1) [81]. Thus, in other studies, rs231775 in CTLA4 gene was not associated with the disease in Korean [82] or Caucasian [80] populations.
Moreover, in a recent review the authors have emphasized other SNPs in genes associated with psoriasis (Table 1) [83]: interferon induced with helicase C domain 1 (IFIH1; rs17716942), late cornified envelope (LCE; rs4085613, rs4845454, rs1886734, rs4112788, rs6701216, and rs4112788), and ring finger protein 114 (RNF114; rs2235617 and rs495337). These genes have also been related with immune system (Table 1): IFIHI with response to viral infections, LCE with epidermal skin barrier function, and RNF114 with T-cell activation. Although, the SNP rs67011216 in LCE gene has been associated with psoriasis in a GWAS study of 223 patients with psoriasis (91 of them with psoriatic arthritis) [42], other authors did not find this association in patients with psoriatic arthritis (n = 1057 cases and n = 5575 controls) [84]. Previously, Zhang et al. (2009) have found an association between rs4112788 in LCE gene and psoriasis in a GWAS performed in Chinese population [85]. A case-control study performed in patients with psoriatic arthritis has found this same association in Caucasian population [86].
In addition, Hébert et al. (2012) supported that the knowledge of risk genes for psoriasis may be useful to predict the response to treatment in patients with this disease [83].
In summary, the literature on the genes involved in immune system that participate in the pathogenesis of psoriasis indicates that IL23R, IL10, TNFα, IL12B, GBP6, IL6, IL13, TNFAIP3, TNIP1, IL1RN, HLA-C, NF-κBIA, APOE, VDR, IFNγ, IL2, IL4, IL15, TNFRSF1B, MCP1, CTLA4, DEFB4, STAT4, IL18, IL19, IL20, IL20RA, ERAP1, IL1B, TRAF3IP2, IL28RA, TYK2, IFIH1, LCE, and ZNF313 play an important role in the development of this disease.
3. Pharmacogenetics of Biological Drugs
3.1. Biological Drugs
The use of agents that block the action of TNFα (infliximab, etanercept, and adalimumab) has shown clear benefits in the treatment of patients with inflammatory diseases such as psoriasis [87]. TNFα induces the production of proinflammatory cytokines such as IL1 and IL6 (Figure 1), which in turn limits leukocyte migration and expression of adhesion molecules by endothelial cells and leukocytes. Neutralization of the biological activity of TNFα leads to an overall reduction in inflammation. Although anti-TNFα therapy is safe and well tolerated, some adverse events have been reported [88].
Advances in knowledge of the metabolic pathways involved in the pathogenesis of psoriasis and related diseases have led to the search for new therapeutic targets and the development of new biological drugs [10]. Such is the case of ustekinumab, a novel human immunoglobulin IgG1κ monoclonal antibody that binds strongly to the p40 subunit shared by IL12 and IL23 (Figure 1). This drug was designed to block the inflammatory cascade of Th1 and Th17 lymphocytes, since the altered behavior of keratinocytes in psoriasis probably results in deregulation of these pathways (Figure 1) [89]. In general, ustekinumab was well tolerated [90].
As mentioned above, psoriasis is mediated by the Th1/Th17 response. New biological therapies—both anti-IL17 agents (ixekizumab and secukinumab) [91, 92] and anti-IL17R agents (brodalumab) [93]—are being developed for the treatment of moderate-to-severe plaque psoriasis (Figure 1). Anti-IL17 drugs are now in phase III trials and may become new alternatives to ustekinumab and anti-TNF therapy [9]. Findings for anti-IL17 and anti-IL17R drugs illustrate the importance of the role of IL17 in the pathogenesis of psoriasis [18, 94].
3.2. Other Treatments of Psoriasis in the Future
Biological drugs are well tolerated and improve the PASI-75 (Psoriasis Area and Severity Index reduction ≥75%) score at week 12 [88, 92, 93, 95, 96]. Their main disadvantages are that injectable administration may cause rejection in some patients. Orally administered alternatives—tofacitinib and apremilast—are being developed (Figure 1).
Tofacitinib is a small JAK1/3 inhibitor molecule that was developed to treat psoriasis and other inflammatory diseases (Figure 1) [97]. The JAK family plays a key role in signal transduction from cytokine receptor in lymphocytes to STAT, which is involved in immune responses (Figure 1) [10, 98].
Apremilast is a PDE4 inhibitor that increases levels of cyclic adenosine monophosphate (cAMP) (Figure 1), which activates the protein kinase A and modulates the cytokines involved in the immune response of psoriasis (decreases TNFα, IL23, and IFNγ and increases IL10) [3]. PDE4 inhibitors cause anti-inflammatory activities [99], such as modulation of the synthesis and release of cytokines and chemokines from immune system cells. Stimulation with TNFα and IL1β can release several mediators: IL8, eotaxin-1, macrophage inflammatory protein 1-α (MIP1α/CCL3), MCP1, and chemokine regulated on activation, normal T cells expressed and secreted (RANTES/CCL5) (Figure 1) [99]. PDE4 inhibitors also suppress the production of inflammatory mediators by Th1 (IL2, IFNγ), Th2 (IL4), and macrophages (TNFα) but increase IL10 synthesis (Figure 1) [99]. Phase II studies have shown an acceptable tolerability and safety profile [100]. Phase III clinical trials of apremilast are ongoing.
Below, we review a selection of pharmacogenetics studies evaluating the efficacy and safety profile of biological drugs.
3.3. Pharmacogenetics
Only two studies have reported the effect of polymorphisms on the response to drugs used to treat psoriasis. In the first, Tejasvi et al. (2012) evaluated associations between two SNPs in TNFAIP3 (rs2230926 and rs610604) and the response to TNF therapy in a cohort from Michigan (n = 433 patients) and a cohort from Toronto (n = 199 patients), both comprising patients with psoriasis and psoriatic arthritis [15]. The SNP rs610604 in TNFAIP3 gene had previously been associated with predisposition to psoriasis and psoriatic arthritis [101]. The authors showed a favorable response to anti-TNF drugs (etanercept, infliximab, and adalimumab) and etanercept in carriers of the G allele of rs610604 in TNFAIP3 in their Michigan cohort (OR = 1.5 and OR = 1.64, resp.) (Table 1). The T-G haplotype of rs2230926-rs610604 (TNFAIP3) was also associated with the response to anti-TNF in this cohort (Table 1). The authors did not find significant differences between rs610604 in TNFAIP3 gene and adalimumab or infliximab analyzed individually or between the SNPs studied and the response to anti-TNF drugs in the Toronto cohort. The study presented the differences in the results between the two cohorts, stating that the reduced size of the Toronto cohort was a limitation of the study [15].
The other study was performed in 80 Greek psoriatic patients (43 women and 37 men) treated with adalimumab, infliximab, and etanercept. The authors analyzed five polymorphisms in three genes: TNFα (rs361525, rs1800629, rs1799724), TNFRSF1A (rs767455), and TNFRSF1B (rs1061622) [14]. Genotyping data revealed an association with response to treatment after 6 months; the patients who achieved a reduction in the PASI score >75% were classified as responders and those with a reduction of ≤50% were classified as nonresponders [14].
Vasilopoulos et al. [14] found an association between a polymorphism in TNFα (CC genotype for rs1799724; P = 0.027) and in TNFRSF1B (TT genotype for rs1061622; P = 0.019) and a better response to anti-TNF treatment (Table 1). The statistical analysis of each agent separately revealed an association between these genotypes and a positive response to etanercept after 6 months of therapy (P = 0.002 and P = 0.001, resp.). However, these SNPs were not associated with a good response to infliximab or adalimumab. The authors explained these differences by the mode of action of biological drugs (etanercept binds to soluble TNFα, and adalimumab and infliximab bind to transmembrane TNFα). The tests of association between the haplotype rs1799724-rs1061622 (TNFα-TNFRSF1B genes) and the response to anti-TNF drugs showed significant differences (P < 0.05) for CT, CG, and TG. It is important to note that Vasilopoulos et al. excluded rs361525 (TNFα), rs1800629 (TNFα), and rs767455 (TNFRSF1A) from the statistical analysis because of a deviation from the Hardy-Weinberg equilibrium [14]. Nevertheless, other authors have reported that a deviation in Hardy-Weinberg equilibrium indicates a real association between genotype and disease [102].
Before treatment of psoriasis can be personalized, more studies should investigate the polymorphisms presented in this review, as well as other polymorphisms and their possible association with drugs used in the treatment of psoriasis. One recent review reported a role for SNPs in psoriasis-related autoimmune diseases (psoriatic arthritis, rheumatoid arthritis, and Crohn's disease) that could play a role in the response to anti-TNF drugs [8].
4. Conclusions
Our review focused only on those polymorphisms associated with the immune system and psoriasis. Current knowledge is limited, and many other SNPs not associated with immune system may be implicated in the development of psoriasis. Larger studies are necessary to obtain a better understanding of this complex disease, the pathways involved in its pathogenesis, and its pharmacogenetic implications in order to develop more effective and safer drugs that can be administered on a personalized basis.
Conflict of Interests
Esteban Daudén has the following conflict of interests: Advisory Board member, consultant, grants, research support, participation in clinical trials, honorarium for speaking, research support, with the following pharmaceutical companies: AbbVie (Abbott), Amgen, Astellas, Centocor Ortho Biotech Inc., Galderma, Glaxo, Janssen-Cilag, Leo Pharma, Novartis, Pfizer, MSD, and Celgene.
Acknowledgments
The authors are grateful to Instituto de Salud Carlos III (FIS PI10/01740) and Fundación Teófilo Hernando for funding this study and to Mr. Thomas O'Boyle for editorial assistance.
References
- 1.Yamamoto M, Imai Y, Sakaguchi Y, Haneda T, Yamanishi K. Serum cytokines correlated with the disease severity of generalized pustular psoriasis. Disease Markers. 2013;34(3):153–161. doi: 10.3233/DMA-120958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Julia A, Tortosa R, Hernanz JM. Risk variants for psoriasis vulgaris in a large case-control collection and association with clinical subphenotypes. Human Molecular Genetics. 2012;21(20):4549–4557. doi: 10.1093/hmg/dds295. [DOI] [PubMed] [Google Scholar]
- 3.Schafer P. Apremilast mechanism of action and application to psoriasis and psoriatic arthritis. Biochemical Pharmacology. 2012;83(12):1583–1590. doi: 10.1016/j.bcp.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Dauden E, Herrera E, Puig L. Validation of a new tool to assess health-related quality of life in psoriasis: the PSO-LIFE questionnaire. Health and Quality of Life Outcomes. 2012;10:p. 56. doi: 10.1186/1477-7525-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang P, Zhao M, Liang G. Whole-genome DNA methylation in skin lesions from patients with psoriasis vulgaris. Journal of Autoimmunity. 2013 doi: 10.1016/j.jaut.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Chandran V, Raychaudhuri SP. Geoepidemiology and environmental factors of psoriasis and psoriatic arthritis. Journal of Autoimmunity. 2010;34(3):J314–J321. doi: 10.1016/j.jaut.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Chandran V. Genetics of psoriasis and psoriatic arthritis. Indian Journal of Dermatology. 2010;55(2):151–156. doi: 10.4103/0019-5154.62751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prieto-Perez R, Cabaleiro T, Dauden E, Abad-Santos F. Gene polymorphisms that can predict response to anti-TNF therapy in patients with psoriasis and related autoimmune diseases. Pharmacogenomics Journal. 2013 doi: 10.1038/tpj.2012.53. [DOI] [PubMed] [Google Scholar]
- 9.Raychaudhuri SP. Role of IL-17 in psoriasis and psoriatic arthritis. Clinical Reviews in Allergy & Immunology. 2012;44(2):183–193. doi: 10.1007/s12016-012-8307-1. [DOI] [PubMed] [Google Scholar]
- 10.Ortiz-Ibanez K, Alsina MM, Munoz-Santos C. Tofacitinib and other kinase inhibitors in the treatment of psoriasis. Actas Dermo-Sifiliográficas. 2013;104(4):304–310. doi: 10.1016/j.adengl.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Vafadari R, Weimar W, Baan CC. Phosphospecific flow cytometry for pharmacodynamic drug monitoring: analysis of the JAK-STAT signaling pathway. Clinica Chimica Acta. 2012;413(17-18):1398–1405. doi: 10.1016/j.cca.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 12.West K. CP-690550, a JAK3 inhibitor as an immunosuppressant for the treatment of rheumatoid arthritis, transplant rejection, psoriasis and other immune-mediated disorders. Current Opinion in Investigational Drugs. 2009;10(5):491–504. [PubMed] [Google Scholar]
- 13.Wojciechowski D, Vincenti F. Targeting JAK3 in kidney transplantation: current status and future options. Current Opinion in Organ Transplantation. 2011;16(6):614–619. doi: 10.1097/MOT.0b013e32834c23ce. [DOI] [PubMed] [Google Scholar]
- 14.Vasilopoulos Y, Manolika M, Zafiriou E, et al. Pharmacogenetic analysis of TNF, TNFRSF1A, and TNFRSF1B gene polymorphisms and prediction of response to anti-TNF therapy in psoriasis patients in the greek population. Molecular Diagnosis and Therapy. 2012;16(1):29–34. doi: 10.1007/BF03256427. [DOI] [PubMed] [Google Scholar]
- 15.Tejasvi T, Stuart PE, Chandran V, et al. TNFAIP3 gene polymorphisms are associated with response to TNF blockade in psoriasis. Journal of Investigative Dermatology. 2012;132(3):593–600. doi: 10.1038/jid.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulski JK, Kenworthy W, Bellgard M, et al. Gene expression profiling of Japanese psoriatic skin reveals an increased activity in molecular stress and immune response signals. Journal of Molecular Medicine. 2005;83(12):964–975. doi: 10.1007/s00109-005-0721-x. [DOI] [PubMed] [Google Scholar]
- 17.Iizuka H, Takahashi H, Honma M, Ishida-Yamamoto A. Unique keratinization process in psoriasis: late differentiation markers are abolished because of the premature cell death. Journal of Dermatology. 2004;31(4):271–276. doi: 10.1111/j.1346-8138.2004.tb00672.x. [DOI] [PubMed] [Google Scholar]
- 18.Krueger JG, Fretzin S, Suarez-Farinas M, et al. IL-17A is essential for cell activation and inflammatory gene circuits in subjects with psoriasis. Journal of Allergy and Clinical Immunology. 2012;130(1):145–154. doi: 10.1016/j.jaci.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohler T, Kruger A, Schneider PM, et al. A TNF-α promoter polymorphism is associated with juvenile onset psoriasis and psoriatic arthritis. Journal of Investigative Dermatology. 1997;109(4):562–565. doi: 10.1111/1523-1747.ep12337469. [DOI] [PubMed] [Google Scholar]
- 20.Mössner R, Kingo K, Kleensang A, et al. Association of TNF -238 and -308 promoter polymorphisms with psoriasis vulgaris and psoriatic arthritis but not with pustulosis palmoplantaris. Journal of Investigative Dermatology. 2005;124(1):282–284. doi: 10.1111/j.0022-202X.2004.23556.x. [DOI] [PubMed] [Google Scholar]
- 21.Louis E, Franchimont D, Piron A, et al. Tumour necrosis factor (TNF) gene polymorphism influences TNF-α production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clinical and Experimental Immunology. 1998;113(3):401–406. doi: 10.1046/j.1365-2249.1998.00662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Wang G, Gao Y, Liu L, Gao T. TNF-α gene promoter −238G>A and −308G>A polymorphisms alter risk of psoriasis vulgaris: a meta-analysis. Journal of Investigative Dermatology. 2007;127(8):1886–1892. doi: 10.1038/sj.jid.5700822. [DOI] [PubMed] [Google Scholar]
- 23.Kaluza W, Reuss E, Grossmann S, et al. Different transcriptional activity and in vitro TNF-α production in psoriasis patients carrying the TNF-α 238A promoter polymorphism. Journal of Investigative Dermatology. 2000;114(6):1180–1183. doi: 10.1046/j.1523-1747.2000.00001.x. [DOI] [PubMed] [Google Scholar]
- 24.Reich K, Mössner R, König IR, Westphal G, Ziegler A, Neumann C. Promoter polymorphisms of the genes encoding tumor necrosis factor-α and interleukin-1β are associated with different subtypes of psoriasis characterized by early and late disease onset. Journal of Investigative Dermatology. 2002;118(1):155–163. doi: 10.1046/j.0022-202x.2001.01642.x. [DOI] [PubMed] [Google Scholar]
- 25.Settin A, Hassan H, El-Baz R, Hassan T. Association of cytokine gene polymorphisms with psoriasis in cases from the Nile Delta of Egypt. Acta Dermatovenerologica Alpina, Pannonica et Adriatica. 2009;18(3):105–112. [PubMed] [Google Scholar]
- 26.Kim T-G, Pyo C-W, Hur S-S, et al. Polymorphisms of tumor necrosis factor (TNF) α and β genes in Korean patients with psoriasis. Archives of Dermatological Research. 2003;295(1):8–13. doi: 10.1007/s00403-003-0392-9. [DOI] [PubMed] [Google Scholar]
- 27.Reich K, Westphal G, Schulz T, et al. Combined analysis of polymorphisms of the tumor necrosis factor-α and interleukin-10 promoter regions and polymorphic xenobiotic metabolizing enzymes in psoriasis. Journal of Investigative Dermatology. 1999;113(2):214–220. doi: 10.1046/j.1523-1747.1999.00654.x. [DOI] [PubMed] [Google Scholar]
- 28.Nedoszytko B, Szczerkowska-Dobosz A, Zabłotna M, Gleń J, Rȩbała K, Roszkiewicz J. Associations of promoter region polymorphisms in the tumour necrosis factor-α gene and early-onset psoriasis vulgaris in a northern Polish population. British Journal of Dermatology. 2007;157(1):165–167. doi: 10.1111/j.1365-2133.2007.07993.x. [DOI] [PubMed] [Google Scholar]
- 29.Arias AI, Giles B, Eiermann TH, Sterry W, Pandey JP. Tumor necrosis factor-alpha gene polymorphism in psoriasis. Experimental and Clinical Immunogenetics. 1997;14(2):118–122. [PubMed] [Google Scholar]
- 30.Johansen C, Vinter H, Soegaard-Madsen L, et al. Preferential inhibition of the mRNA expression of p38 mitogen-activated protein kinase regulated cytokines in psoriatic skin by anti-TNFα therapy. British Journal of Dermatology. 2010;163(6):1194–1204. doi: 10.1111/j.1365-2133.2010.10036.x. [DOI] [PubMed] [Google Scholar]
- 31.Toussirot É. The IL23/Th17 pathway as a therapeutic target in chronic inflammatory diseases. Inflammation and Allergy. 2012;11(2):159–168. doi: 10.2174/187152812800392805. [DOI] [PubMed] [Google Scholar]
- 32.Kurzeja M, Rudnicka L, Olszewska M. New interleukin-23 pathway inhibitors in dermatology: ustekinumab, briakinumab, and secukinumab. American Journal of Clinical Dermatology. 2011;12(2):113–125. doi: 10.2165/11538950-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Begovich AB. Unraveling the genetics of complex diseases: susceptibility genes for rheumatoid arthritis and psoriasis. Seminars in Immunology. 2009;21(6):318–327. doi: 10.1016/j.smim.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Garcia VE, Chang M, Brandon R, et al. Detailed genetic characterization of the interleukin-23 receptor in psoriasis. Genes and Immunity. 2008;9(6):546–555. doi: 10.1038/gene.2008.55. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X-J. Enlightenment from genome-wide association study to genetics of psoriasis. Journal of Zhejiang University. 2009;38(4):333–337. [PubMed] [Google Scholar]
- 36.Nair RP, Ruether A, Stuart PE, et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. Journal of Investigative Dermatology. 2008;128(7):1653–1661. doi: 10.1038/sj.jid.5701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cargill M, Schrodi SJ, Chang M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. American Journal of Human Genetics. 2007;80(2):273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nair RP, Stuart PE, Kullavanijaya P, et al. Genetic evidence for involvement of the IL23 pathway in Thai psoriatics. Archives of Dermatological Research. 2010;302(2):139–143. doi: 10.1007/s00403-009-0986-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y, Lu Z, Chen Y, Xue F, Chen X, Zheng J. Replication of association between interleukin-23 receptor (IL-23R) and its ligand (IL-12B) polymorphisms and psoriasis in the Chinese Han population. Human Immunology. 2010;71(12):1255–1258. doi: 10.1016/j.humimm.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 40.Capon F, Di Meglio P, Szaub J, et al. Sequence variants in the genes for the interleukin-23 receptor (IL23R) and its ligand (IL12B) confer protection against psoriasis. Human Genetics. 2007;122(2):201–206. doi: 10.1007/s00439-007-0397-0. [DOI] [PubMed] [Google Scholar]
- 41.Smith RL, Warren RB, Eyre S, et al. Polymorphisms in the IL-12β and IL-23R genes are associated with psoriasis of early onset in a UK cohort. Journal of Investigative Dermatology. 2008;128(5):1325–1327. doi: 10.1038/sj.jid.5701140. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Helms C, Liao W, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genetics. 2008;4(3) doi: 10.1371/journal.pgen.1000041.e1000041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hüffmeier U, Lascorz J, Böhm B, et al. Genetic variants of the IL-23R pathway: association with psoriatic arthritis and psoriasis vulgaris, but no specific risk factor for arthritis. Journal of Investigative Dermatology. 2009;129(2):355–358. doi: 10.1038/jid.2008.233. [DOI] [PubMed] [Google Scholar]
- 44.Nair RP, Duffin KC, Helms C, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nature Genetics. 2009;41(2):199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellinghaus E, Ellinghaus D, Stuart PE, et al. Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nature Genetics. 2010;42(11):991–995. doi: 10.1038/ng.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsunemi Y, Saeki H, Nakamura K, et al. Interleukin-12 p40 gene (IL12B) 3′-untranslated region polymorphism is associated with susceptibility to atopic dermatitis and psoriasis vulgaris. Journal of Dermatological Science. 2002;30(2):161–166. doi: 10.1016/s0923-1811(02)00072-5. [DOI] [PubMed] [Google Scholar]
- 47.Musone SL, Taylor KE, Nititham J, et al. Sequencing of TNFAIP3 and association of variants with multiple autoimmune diseases. Genes and Immunity. 2011;12(3):176–182. doi: 10.1038/gene.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lodolce JP, Kolodziej LE, Rhee L, et al. African-derived genetic polymorphisms in TNFAIP3 mediate risk for autoimmunity. Journal of Immunology. 2010;184(12):7001–7009. doi: 10.4049/jimmunol.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hüffmeier U, Uebe S, Ekici AB, et al. Comon variants at TRAF3IP2 are asociated with susceptibility to psoriatic arthritis and psoriasis. Nature Genetics. 2010;42(11):996–999. doi: 10.1038/ng.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strange A, Capon F, Spencer CCA, et al. A genome-wide asociation study identifies new psoriasis susceptibility loci and an interaction betwEn HLA-C and ERAP1. Nature Genetics. 2010;42(11):985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng B-J, Sun L-D, Soltani-Arabshahi R, et al. Multiple loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genetics. 2009;5(8) doi: 10.1371/journal.pgen.1000606.e1000606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun LD, Cheng H, Wang ZX. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nature Genetics. 2010;42(11):1005–1009. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim Y-K, Pyo C-W, Choi H-B, Kim S-Y, Kim T-Y, Kim T-G. Associations of IL-2 and IL-4 gene polymorphisms with psoriasis in the Korean population. Journal of Dermatological Science. 2007;48(2):133–139. doi: 10.1016/j.jdermsci.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 54.Baran W, Szepietowski JC, Mazur G, Baran E. IFN-γ promoter gene polymorphism in psoriasis vulgaris. Biomarkers. 2008;13(1):52–58. doi: 10.1080/13547500701610273. [DOI] [PubMed] [Google Scholar]
- 55.Kanda N, Masahiro N, Tada Y, Ishikawa T, Sato S, Watanabe S. Human β-defensin-2 enhances IFN-γ and IL-10 production and suppresses IL-17 production in T cells. Journal of Leukocyte Biology. 2011;89(6):935–944. doi: 10.1189/jlb.0111004. [DOI] [PubMed] [Google Scholar]
- 56.Hollox EJ, Huffmeier U, Zeeuwen PLJM, et al. Psoriasis is associated with increased β-defensin genomic copy number. Nature Genetics. 2008;40(1):23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Web page National Center for Biotecnology Information (NCBI), 2013, http://www.ncbi.nlm.nih.gov/gene.
- 58.Kato T, Tsunemi Y, Saeki H, et al. Interferon-18 gene polymorphism -137 G/C is associated with susceptibility to psoriasis vulgaris but not with atopic dermatitis in Japanese patients. Journal of Dermatological Science. 2009;53(2):162–163. doi: 10.1016/j.jdermsci.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 59.Baran W, Szepietowski JC, Mazur G, Baran E. IL-6 and IL-10 promoter gene polymorphisms in psoriasis vulgaris. Acta Dermato-Venereologica. 2008;88(2):113–116. doi: 10.2340/00015555-0427. [DOI] [PubMed] [Google Scholar]
- 60.Craven NM, Jackson CW, Kirby B, et al. Cytokine gene polymorphisms in psoriasis. British Journal of Dermatology. 2001;144(4):849–853. doi: 10.1046/j.1365-2133.2001.04143.x. [DOI] [PubMed] [Google Scholar]
- 61.Chang M, Li Y, Yan C, et al. Variants in the 5q31 cytokine gene cluster are associated with psoriasis. Genes and Immunity. 2008;9(2):176–181. doi: 10.1038/sj.gene.6364451. [DOI] [PubMed] [Google Scholar]
- 62.Duffin KC, Freeny IC, Schrodi SJ, et al. Association between IL13 polymorphisms and psoriatic arthritis is modified by smoking. The Journal of investigative dermatology. 2009;129(12):2777–2783. doi: 10.1038/jid.2009.169. [DOI] [PubMed] [Google Scholar]
- 63.Bowes J, Eyre S, Flynn E, et al. Evidence to support IL-13 as a risk locus for psoriatic arthritis but not psoriasis vulgaris. Annals of the Rheumatic Diseases. 2011;70(6):1016–1019. doi: 10.1136/ard.2010.143123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kõks S, Kingo K, Rätsep R, Karelson M, Silm H, Vasar E. Combined haplotype analysis of the interleukin-19 and -20 genes: relationship to plaque-type psoriasis. Genes and Immunity. 2004;5(8):662–667. doi: 10.1038/sj.gene.6364141. [DOI] [PubMed] [Google Scholar]
- 65.Chen X-Y, Jin L-W, Chen Y-W, et al. The association between the IL-20 -1723C→G allele on the 1q chromosome and psoriasis triggered or exacerbated by an upper respiratory tract infection in the Chinese Han population. Dermatology. 2011;222(1):24–30. doi: 10.1159/000320772. [DOI] [PubMed] [Google Scholar]
- 66.Kingo K, Kõks S, Nikopensius T, Silm H, Vasar E. Polymorphisms in the interleukin-20 gene: relationships to plaque-type psoriasis. Genes and Immunity. 2004;5(2):117–121. doi: 10.1038/sj.gene.6364046. [DOI] [PubMed] [Google Scholar]
- 67.Kingo K, Mössner R, Traks T, et al. Further association analysis of chr 6q22-24 suggests a role of IL-20RA polymorphisms in psoriasis. Journal of Dermatological Science. 2010;57(1):71–73. doi: 10.1016/j.jdermsci.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 68.Kõks S, Kingo K, Vabrit K, et al. Possible relations between the polymorphisms of the cytokines IL-19, IL-20 and IL-24 and plaque-type psoriasis. Genes and Immunity. 2005;6(5):407–415. doi: 10.1038/sj.gene.6364216. [DOI] [PubMed] [Google Scholar]
- 69.Zhang X-J, Yan K-L, Wang Z-M, et al. Polymorphisms in interleukin-15 gene on chromosome 4q31.2 are associated with psoriasis vulgaris in Chinese population. Journal of Investigative Dermatology. 2007;127(11):2544–2551. doi: 10.1038/sj.jid.5700896. [DOI] [PubMed] [Google Scholar]
- 70.Smith RL, Eyre S, Warren RB, Young HS, Griffiths CEM, Worthington J. No association between polymorphisms in the interleukin-15 gene and early-onset psoriasis in a UK cohort suggests heterogeneity for this susceptibility locus identified in Chinese psoriasis patients. Journal of Investigative Dermatology. 2008;128(12):2904–2905. doi: 10.1038/jid.2008.148. [DOI] [PubMed] [Google Scholar]
- 71.Wang L, Yang L, Gao L, Gao TW, Li W, Liu YF. A functional promoter polymorphism in monocyte chemoattractant protein-1 is associated with psoriasis. International Journal of Immunogenetics. 2008;35(1):45–49. doi: 10.1111/j.1744-313X.2007.00734.x. [DOI] [PubMed] [Google Scholar]
- 72.Zervou MI, Goulielmos GN, Castro-Giner F, Tosca AD, Krueger-Krasagakis S. STAT4 gene polymorphism is associated with psoriasis in the genetically homogeneous population of Crete, Greece. Human Immunology. 2009;70(9):738–741. doi: 10.1016/j.humimm.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 73.Karpouzis A, Caridha R, Tripsianis G, Michailidis C, Martinis G, Veletza SV. Apolipoprotein e gene polymorphism in psoriasis. Archives of Dermatological Research. 2009;301(6):405–410. doi: 10.1007/s00403-009-0968-0. [DOI] [PubMed] [Google Scholar]
- 74.Coto-Segura P, Coto E, Alvarez V, et al. Apolipoprotein ε4 allele is associated with psoriasis severity. Archives of Dermatological Research. 2010;302(2):145–149. doi: 10.1007/s00403-009-1002-2. [DOI] [PubMed] [Google Scholar]
- 75.Campalani E, Allen MH, Fairhurst D, et al. Apolipoprotein E gene polymorphisms are associated with psoriasis but do not determine disease response to acitretin. British Journal of Dermatology. 2006;154(2):345–352. doi: 10.1111/j.1365-2133.2005.06950.x. [DOI] [PubMed] [Google Scholar]
- 76.Halsall JA, Osborne JE, Pringle JH, Hutchinson PE. Vitamin D receptor gene polymorphisms, particularly the novel A-1012G promoter polymorphism, are associated with vitamin D3 responsiveness and non-familial susceptibility in psoriasis. Pharmacogenetics and Genomics. 2005;15(5):349–355. doi: 10.1097/01213011-200505000-00011. [DOI] [PubMed] [Google Scholar]
- 77.Zuel-Fakkar NM, Kamel MM, Asaad MK, Mahran MZ, Shehab AA. A study of ApaI and TaqI genotypes of the vitamin D receptor in Egyptian patients with psoriasis. Clinical and Experimental Dermatology. 2011;36(4):355–359. doi: 10.1111/j.1365-2230.2010.03970.x. [DOI] [PubMed] [Google Scholar]
- 78.Rucevic I, Barisic-Drusko V, Glavas-Obrovac L, Stefanic M. Vitamin D endocrine system and psoriasis vulgaris—review of the literature. Acta Dermatovenerologica Croatica. 2009;17(3):187–192. [PubMed] [Google Scholar]
- 79.Dayangac-Erden D, Karaduman A, Erdem-Yurter H. Polymorphisms of vitamin D receptor gene in Turkish familial psoriasis patients. Archives of Dermatological Research. 2007;299(10):487–491. doi: 10.1007/s00403-007-0782-5. [DOI] [PubMed] [Google Scholar]
- 80.Łuszczek W, Kubicka W, Jasek M, et al. CTLA-4 gene polymorphisms and natural soluble CTLA-4 protein in psoriasis vulgaris. International Journal of Immunogenetics. 2006;33(3):217–224. doi: 10.1111/j.1744-313X.2006.00600.x. [DOI] [PubMed] [Google Scholar]
- 81.Łuszczek W, Majorczyk E, Nockowski P, et al. Distribution of the CTLA-4 single nucleotide polymorphisms CT60G>A and +49A>G in psoriasis vulgaris patients and control individuals from a Polish Caucasian population. International Journal of Immunogenetics. 2008;35(1):51–55. doi: 10.1111/j.1744-313X.2007.00736.x. [DOI] [PubMed] [Google Scholar]
- 82.Kim Y-K, Pyo C-W, Hur S-S, Kim T-Y, Kim T-G. No associations of CTLA-4 and ICAM-1 polymorphisms with psoriasis in the Korean population. Journal of Dermatological Science. 2003;33(1):75–77. doi: 10.1016/s0923-1811(03)00159-2. [DOI] [PubMed] [Google Scholar]
- 83.Hébert HL, Ali FR, Bowes J, Griffiths CEM, Barton A, Warren RB. Genetic susceptibility to psoriasis and psoriatic arthritis: implications for therapy. British Journal of Dermatology. 2012;166(3):474–482. doi: 10.1111/j.1365-2133.2011.10712.x. [DOI] [PubMed] [Google Scholar]
- 84.Bowes J, Flynn E, Ho P, et al. Variants in linkage disequilibrium with the late cornified envelope gene cluster deletion are associated with susceptibility to psoriatic arthritis. Annals of the Rheumatic Diseases. 2010;69(12):2199–2203. doi: 10.1136/ard.2010.130575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang X-J, Huang W, Yang S, et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nature Genetics. 2009;41(2):205–210. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- 86.Docampo E, Giardina E, Riveira-Muñoz E, et al. Deletion of LCE3C and LCE3B is a susceptibility factor for psoriatic arthritis: a study in Spanish and Italian populations and meta-analysis. Arthritis and Rheumatism. 2011;63(7):1860–1865. doi: 10.1002/art.30340. [DOI] [PubMed] [Google Scholar]
- 87.Kircik LH, Del Rosso JQ. Anti-TNF agents for the treatment of psoriasis. Journal of Drugs in Dermatology. 2009;8(6):546–559. [PubMed] [Google Scholar]
- 88.Sivamani RK, Correa G, Ono Y, Bowen MP, Raychaudhuri SP, Maverakis E. Biological therapy of psoriasis. Indian Journal of Dermatology. 2010;55(2):161–170. doi: 10.4103/0019-5154.62754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brand S. Crohn’s disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn’s disease. Gut. 2009;58(8):1152–1167. doi: 10.1136/gut.2008.163667. [DOI] [PubMed] [Google Scholar]
- 90.Gandhi M, Alwawi E, Gordon KB. Anti-p40 Antibodies Ustekinumab and Briakinumab: blockade of Interleukin-12 and Interleukin-23 in the Treatment of Psoriasis. Seminars in Cutaneous Medicine and Surgery. 2010;29(1):48–52. doi: 10.1016/j.sder.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 91.Wu JJ. Anti-interleukin-17 monoclonal antibody ixekizumab in psoriasis. New England Journal of Medicine. 367(3):274–275. doi: 10.1056/NEJMc1205835. [DOI] [PubMed] [Google Scholar]
- 92.Rich P, Sigurgeirsson B, Thaci D, et al. Secukinumab induction and maintenance therapy in moderate-to-severe plaque psoriasis: a randomized, double-blind, placebo-controlled, phase II regimen-finding study. British Journal of Dermatology. 2013;168(2):402–411. doi: 10.1111/bjd.12112. [DOI] [PubMed] [Google Scholar]
- 93.Papp KA, Leonardi C, Menter A, et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. New England Journal of Medicine. 2012;366(13):1181–1189. doi: 10.1056/NEJMoa1109017. [DOI] [PubMed] [Google Scholar]
- 94.Patel DD, Lee DM, Kolbinger F, Antoni C. Effect of IL-17A blockade with secukinumab in autoimmune diseases. Annals of the Rheumatic Diseases. 2012 doi: 10.1136/annrheumdis-2012-202371. [DOI] [PubMed] [Google Scholar]
- 95.Leonardi C, Matheson R, Zachariae C, et al. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. New England Journal of Medicine. 2012;366(13):1190–1191. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 96.Papp KA, Langley RG, Sigurgeirsson B, et al. Efficacy and safety of secukinumab in the treatment of moderate-to-severe plaque psoriasis: a randomized, double-blind, placebo-controlled phase II dose-ranging study. British Journal of Dermatology. 2013;168(2):412–421. doi: 10.1111/bjd.12110. [DOI] [PubMed] [Google Scholar]
- 97.Ports WC, Khan S, Lan S, et al. A randomised Phase 2a efficacy and safety trial of the topical Janus kinase inhibitor tofacitinib in the treatment of chronic plaque psoriasis. British Journal of Dermatology. 2013 doi: 10.1111/bjd.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Menet CJ, Rompaey LV, Geney R. Advances in the discovery of selective JAK inhibitors. Progress in Medicinal Chemistry. 2013;52:153–223. doi: 10.1016/B978-0-444-62652-3.00004-1. [DOI] [PubMed] [Google Scholar]
- 99.Sanz MJ, Cortijo J, Morcillo EJ. PDE4 inhibitors as new anti-inflammatory drugs: effects on cell trafficking and cell adhesion molecules expression. Pharmacology and Therapeutics. 2005;106(3):269–297. doi: 10.1016/j.pharmthera.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 100.Papp K, Cather JC, Rosoph L, et al. Efficacy of apremilast in the treatment of moderate to severe psoriasis: a randomised controlled trial. Lancet. 2012;380(9843):738–746. doi: 10.1016/S0140-6736(12)60642-4. [DOI] [PubMed] [Google Scholar]
- 101.Bowes J, Barton A. The genetics of psoriatic arthritis: lessons from genome-wide association studies. Discovery Medicine. 2010;10(52):177–183. [PubMed] [Google Scholar]
- 102.Soriguer F, Morcillo S. Qué hacer cuando en los estudios de epidemiología biomolecular la distribución genotípica no se ajusta al equilibrio de Hardy-Weinberg. Endocrinología y Nutrición. 2007;54(3):169–173. [Google Scholar]
- 103.Zheng H-F, Zuo X-B, Lu W-S, et al. Variants in MHC, LCE and IL12B have epistatic effects on psoriasis risk in Chinese population. Journal of Dermatological Science. 2011;61(2):124–128. doi: 10.1016/j.jdermsci.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 104.Zheng H-F, Zhang C, Sun L-D, et al. A single nucleotide polymorphism of MHC region is associated with subphenotypes of Psoriasis in Chinese population. Journal of Dermatological Science. 2010;59(1):50–52. doi: 10.1016/j.jdermsci.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 105.Capon F, Bijlmakers M-J, Wolf N, et al. Identification of ZNF313/RNF114 as a novel psoriasis susceptibility gene. Human Molecular Genetics. 2008;17(13):1938–1945. doi: 10.1093/hmg/ddn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Web page Hapmap, 2013, http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap3r3_B36/#search.
- 107. Web page Alfred, 2013, http://alfred.med.yale.edu/


