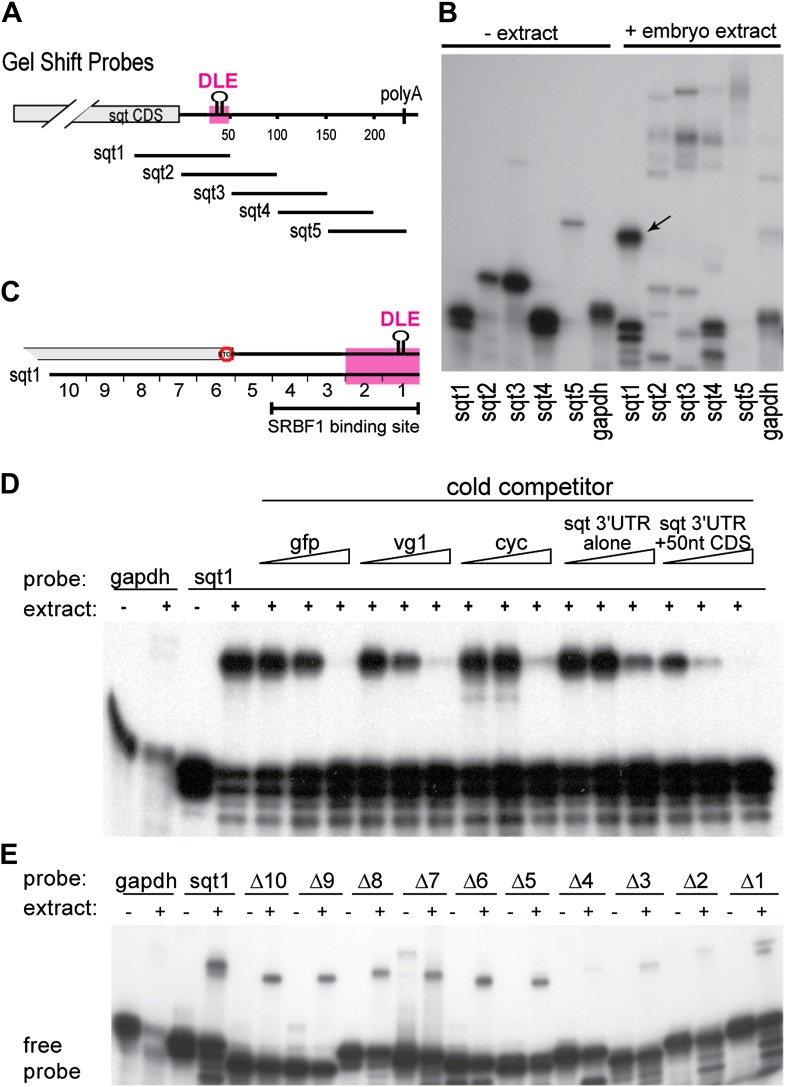
Figure 1. SRBF1 binds the sqt Dorsal Localization Element (DLE).
(A) Schematic of overlapping 100 nucleotide radioactive RNA gel-shift probes spanning the sqt 3′UTR. Position of DLE is highlighted in magenta. (B) Autoradiogram showing sqt 3′UTR probes incubated with embryo extract. Several binding activities were observed on the various probes. The ‘sqt RNA Binding Factor 1’ (SRBF1; black arrow) shift, is detected on the DLE-containing sqt1 probe, and not on other probes. (C) Schematic showing the SRBF1 binding site. sqt DLE is highlighted in magenta and the red octagon indicates the stop codon. (D) Competition gel-shift assay shows that SRBF1 binds specifically to sqt RNA. The sqt 3′UTR with 50 nucleotides of coding sequence competes more strongly than control gfp, vg1 or cyclops (cyc) RNA for binding to sqt1 probe. Triangles represent 4-fold increases (from 5 ng to 80 ng) of cold competitor RNA. Thus, SRBF1 preferentially binds DLE-sequences. (E) The SRBF1 binding site overlaps the DLE. RNA gel-shifts were performed with the sqt1 10 nt deletion series.
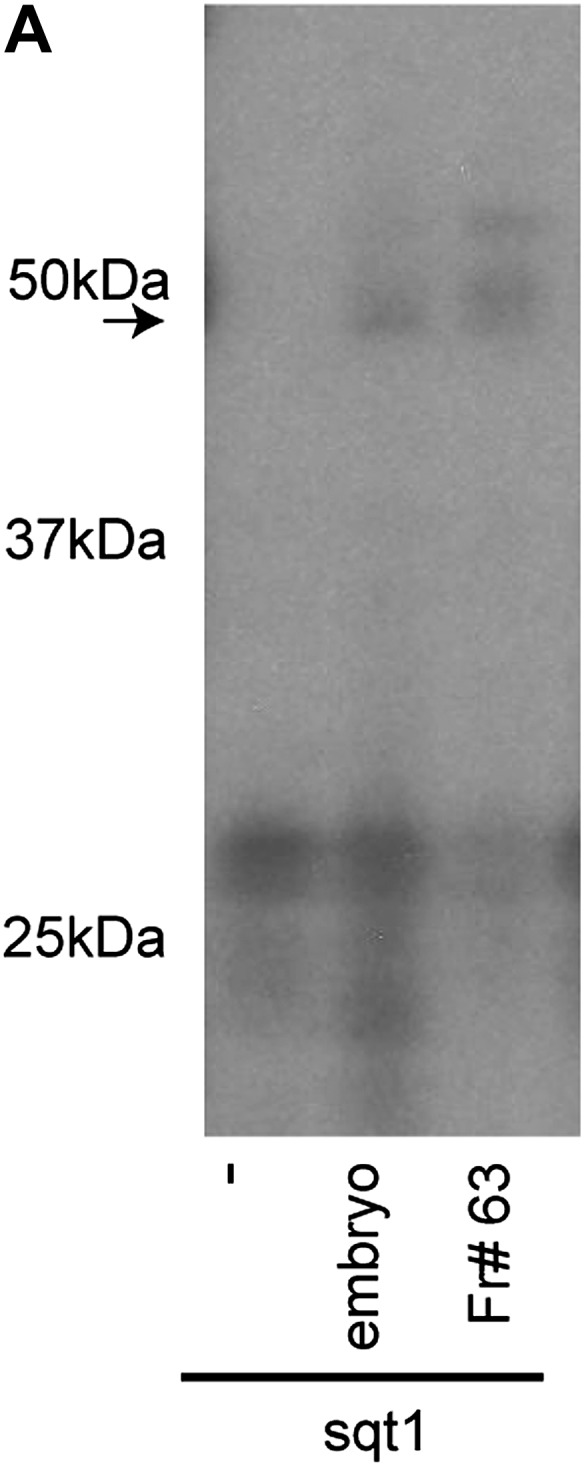
Figure 1—figure supplement 1. SRBF1 is an ∼50 kDa protein.