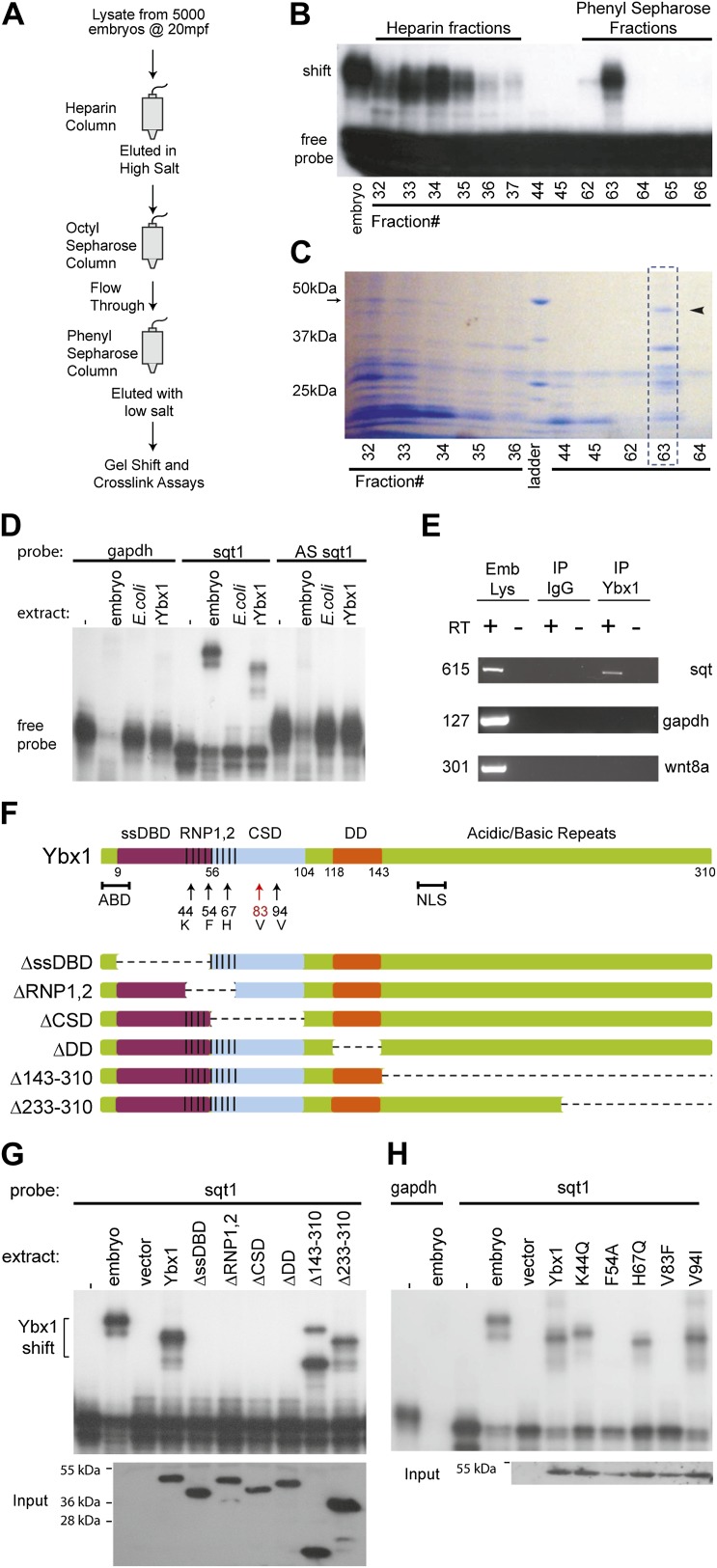
Figure 2. SRBF1 contains the nucleic acid binding protein Ybx1.
(A) Extracts from 5000 embryos collected at 20 mpf were sequentially fractionated on multiple chromatography columns, until SRBF1 was partially pure. At each stage, fractions containing SRBF1 activity were pooled, and loaded onto the next column for further purification. (B) A representative native PAGE gel showing SRBF1 purification. Gel-shift analysis of fractions from the heparin and phenyl sepharose columns show SRBF1 activity in fractions 32–37 from heparin and fractions 62–63 from the phenyl sepharose columns. Fractions 33–35 were pooled and added to the hydrophobic columns. Fraction 63 from the phenyl sepharose column contains partially purified SRBF1. (C) A Coomassie-blue stained SDS-PAGE gel of the fractions in B show a ∼48 kDa band that co-fractionates with SRBF1 (black arrowhead in fraction#63). The 48 kDa band from fraction#63 was excised, sequenced by mass spectrometry, and found to contain Ybx1 peptides. (D) Gel-shift analysis shows recombinant Ybx1 (rYbx1), similar to embryonic SRBF1, binds to sqt1, but not to control gapdh or antisense sqt1 probes. (E) Ybx1 binds sqt RNA in vivo. RT-PCR shows sqt RNA but not control gapdh or wnt8a RNA in RNA-IP with αYbx1 antibodies. Control IgG antibodies do not show any RT-PCR product. RT-PCR from whole embryo lysates is the positive control. PCR product sizes are indicated on the left. (F) Schematic diagram showing domain structure of wild-type and mutant Ybx1 proteins. The position of amino acid substitutions is indicated by arrows (V83 in red and all other residues in black). Deletions are indicated by dashed lines. The actin binding domain (ABD), single stranded DNA-binding domain (ssDBD; magenta), RNA-binding domains 1 and 2 (RNP1,2; hashed black lines), Cold shock domain (CSD, blue), dimerization domain (DD; orange), and nuclear localization sequence (NLS) are shown; numbers indicate the amino acid residue. (G) Domain analysis of Ybx1. The nucleic acid binding domain (ssDBD, magenta bar in F; CSD, blue bar in F; RNP1,2, hashed lines in F) is required for binding to sqt1, as is the dimerization domain (DD, orange bar in F). In contrast, the C-terminus of Ybx1 (Δ143–310 and Δ233–310) is dispensable for binding to sqt1. A western blot with α-His tag antibodies shows expression of the mutant Ybx1 proteins. (H) Point mutations in Ybx1 identify key amino acid residues that confer sqt RNA binding. K44, F54, and H67 are expected to contact RNA based upon NMR structure prediction. F54A abolishes binding, whereas H67Q does not affect binding at the protein concentrations shown. V83F abolishes sqt1 binding, whereas V94I binds sqt1. Western blot with α-His tag antibodies shows expression of mutant Ybx1 proteins.