Abstract
Notch signaling is among the oldest of known Metazoan signaling pathways and is used in a multitude of developmental contexts to effect cellular differentiation, specification and the maintenance of stem cell state. Here we report the isolation and expression of the canonical Notch signaling pathway in the early branching metazoan Nematostella vectensis (Anthozoa, Cnidaria) during embryonic and larval development. We have used pharmacological treatment, morpholino knockdown, and dominant negative misexpression experiments to demonstrate that Notch signaling acts to mediate cnidogenesis, the development of cnidarian-specific neural effecter cells. Notch signaling often results in the transcriptional activation of NvHes genes, a conserved family of bHLH transcription factors. A loss of Notch signaling through use of pharmacological inhibition or knock-down of the Notch effecter gene Suppressor of Hairless Su(H) similarly results in a loss of cnidocyte cell fate. We also provide evidence that Notch signaling is responsible for certain aspects of neurogenesis in developing N. vectensis planula in which disruption of Notch cleavage via the pharmacological agent DAPT results in increased expression of neural marker genes in vivo. This data suggests that Notch signaling acting on components of the developing nervous system is an ancient role of this pathway. The shared requirement of Notch signaling for the development of both cnidocytes and neurons further supports the hypothesis that cnidocytes and neurons share common origins as multifunctional sensory-effecter cells.
Keywords: cnidarian, notch evolution, evo devo, neurogenesis, cnidogenesis
Introduction
The Notch intracellular signaling pathway is unusual in its requirement of cell-cell contact for activity of the membrane bound Notch receptor and ligand as well as for its direct mechanism of signal transduction through endocytosis of a portion of the receptor without amplification through second-messenger cascades (Kopan and Ilagan, 2009). Notch receptors are characterized by an extracellular domain containing both ligand recognition as well as negative control regions that mediate receptor cleavage and the subsequent activation of the pathway (Kopan and Ilagan, 2009; Le Borgne et al., 2005). Notch activation is triggered by binding of a DSL domain-bearing ligand, that like Notch, must be endocytosed for proper activity of the pathway. Once cleaved, the intracellular domain of Notch acts directly through conserved protein domains as a co-transcriptional activator with suppressor of hairless (Su(H)/RBPjK) on downstream targets. These targets include hairy and enhancer of split (Hes), and enhancer of split (E)spl as well as a growing number of reported potential targets, many of which are in themselves transcription factors. Notch, these downstream targets, and ligands of Notch form the “canonical Notch signaling pathway”(Kopan and Ilagan, 2009; Schweisguth, 1995). While canonical Notch signaling may represent the dominant mode of pathway activity, new modes of action have been discovered in which Notch may interact with other signaling pathways such as NFkB (Ang and Tergaonkar, 2007), or newly described families of ligands (Chen and Greenwald, 2004), that act independently of Su(H)/RBPjK (Arias, 2002; Hori et al., 2004; Matsuno et al., 1997) . Understanding of the role of Su(H)/RBPjK protein, the main co-transcriptional activator for the Notch intracellular domain, has improved in recent years and has revealed additional roles for Su(H) as a Notch-independent transcription factor (Barolo et al., 2000; Koelzer and Klein, 2003), as a transcriptional repressor in the absence of Notch signaling, and as an indirect inducer of epigenetic modifications (Kopan and Ilagan, 2009).
Notch signaling can mediate a number of cell-cell signaling events in a broad range of developmental roles. One of the best-documented roles of Notch is in the development of neural precursors from epithelial tissues by way of lateral inhibition in embryonic Drosophila neurogenesis (Heitzler et al., 1996; Simpson, 1990). Notch can also function through inductive signaling such as that described for the specification of the AB cell lineage in the four cell C. elegans embryo wherein the P2 cell, expressing the apx-1/delta ligand signals to the glp-1/Notch-bearing AB cell lineage (Mickey et al., 1996). Notch also maintains roles in adult vertebrate neurogenesis by inductive and repressive signaling (Louvi and Artavanis-Tsakonas, 2006). Recent studies have identified Notch as a key regulatory component of both the vertebrate segmentation clock (Lewis et al., 2009; Ozbudak and Pourquie, 2008) and of segmentation in arthropods (Chipman and Akam, 2008). The deployment of Notch as mediator of many developmental processes as well as its role in the development of taxonomically restricted morphological novelties, such as secondary mesenchyme cells and their derivatives in sea urchins (Sherwood and McClay, 1999) further supports the view of Notch as a flexible pathway that has been co-opted repeatedly over evolutionary time. Comparative developmental approaches are required to determine the most ancient roles of the Notch pathway, particularly with respect to the earliest appearing animal phyla.
Cnidarians are an early branching basal metazoan group with a relatively simple body plan (Darling et al., 2005). Anthozoan cnidarians such as Nematostella vectensis, the starlet sea aneomone, are diploblastic with only a single gut opening and two main body axes, the oral-aboral and directive axes (Finnerty et al., 2004). Despite their apparent morphological simplicity, cnidarians have been shown to have considerable genomic complexity and possess nearly complete repertoires of all major metazoan signaling pathways (Putnam et al., 2007). This enigmatic contrast between molecular complexity and morphological readout has prompted a number of studies on the role of developmental signaling pathways in these ancient animals (Lee et al., 2006; Matus et al., 2007b). Surprisingly, a number of parallels between Cnidarian and Bilaterian development have emerged. It has been demonstrated that the Wnt/Beta-catenin pathway plays a functional role in establishing the embryonic oral-aboral axis, the major body axis, and in the specification of the bi-functional endomesodermal layer, much as it does in deuterostome embryos (Lee et al., 2007; Wikramanayake et al., 2003). In addition, the FGF signaling pathway has been implicated in the formation of the major larval neural structure, the apical tuft (Rentzsch et al., 2008).
The presence of the Notch signaling pathway in basal metazoans such as N. vectensis is particularly fascinating considering its nearly universal requirement throughout Bilateria in mediating cell-cell communication in extremely varied developmental contexts. This requirement for local signaling to specify defined sub-populations of cells makes the Notch pathway a particularly important candidate for study in early cnidarian embryogenesis. A lack of cnidarian cell-type specific markers has historically proved a limiting factor in assaying morphological readout of molecular events, however recent work with newly available genomic resources has spurred the development of such reagents (Marlow et al., 2009; Matus et al., 2007a) and leaves N. vectensis development poised for such functional investigations into local signaling events mediated by the Notch pathway.
Notch signaling is hypothesized to be an ancient metazoan feature as all metazoans thus far examined, including sponges, have at least one Notch receptor and one DSL-bearing ligand but we have little information regarding their function in these lineages (Gazave et al., 2009). In sponges, Notch, delta and a single proneural bHLH gene are suggested to mediate development of epithelial globular cells, described as ancient sensory cells (Richards et al., 2008). When ectopically expressed in Xenopus and Drosophila, the sponge AmqbHLH1 gene induces a neurogenic phenotype (Richards et al., 2008). In addition, AmqbHLH1 can induce delta expression and is inhibited by a constiuitively active form of Notch in Xenopus indicating that the molecular mechanism of the Notch signaling pathway is also conserved (Richards et al., 2008). We do not know, however, what the functional role of Notch signaling in sponges is within the context of the developing embryo or what relationship the globular cells have with the nervous systems of cnidarians and bilaterians.
Eumetazoans such as the cnidarian N. vectensis and bilaterians have Notch and two DSL containing ligands, delta and serrate/jagged, indicating that an ancient duplication of DSL-bearing ligands occurred prior to the divergence of cnidarians and bilaterians (Gazave et al., 2009). Downstream targets of Notch signaling, the Hes/(E)spl genes have been identified in Nematostella as well (Simionato et al., 2007). Initial studies in the Cnidaria indicate that Notch signaling via cleavage and translocation of the NICD have been reported in adults of the model hydrozoan Hydra. Studies on adult hydra indicate that Notch has a role in germ and cnidocyte cell differentiation, however, these treatments do not appear to impact other aspects of neurogenesis (Kasbauer et al., 2007). Recent studies on budding hydrozoans implicate a role for Notch in boundary formation in the forming bud as well and the capacity for HyNotch to regulate the expression of HyHes by interacting with its promoter region (Munder et al., 2010). We have chosen to examine the role of the Notch pathway in the cnidarian N. vectensis to better understand its function during development, which is relatively inaccessible in Hydra. In addition, anthozoans such as N. vectensis have a dramatically different mode of neurogenesis than that found in the Hydrozoa (Galliot et al., 2009; Marlow et al., 2009) and might have therefore retained a role for Notch signaling in neurogenesis, a role observed in virtually all bilaterian lineages.
Methods
Identification of Notch pathway genes and probe synthesis for in situ hybridization
Genes encoding predicted open reading frames for notch and delta transcripts were identified by performing blastp and tblastn searches of the assembled N. vectensis genome (Joint Genome Institute http://genome.jgi-psf.org/Nemve1/Nemve1.home.html) using bilaterian NOD and DSL domains. Predicted open reading frames for hairy and enhancer of split (hes) were identified through blastp and tblastn searches of the N. vectensis genome using hes genes from vertebrates and enhancer of split (e(spl)) genes from Drosophila. Several DSL-containing genes as well as many hes-like genes were identified and additional 5′ and 3′ sequence was isolated using rapid amplification of cDNA ends with RACE PCR (Smart Race cDNA amplification kit, Clontech, Inc.). Sequences were cloned into the p-GEM T Easy vector (Promega, Inc.). Resulting sequences were aligned using Sequencher 4.1 and 4.6 (Gene Codes Corporation, Inc.) and prediction of conserved protein domains was performed using the SMART protein (Ponting et al., 1999), the Conserved Domain Database (Geer et al., 2002), and NLStradamus (Nguyen Ba et al., 2009). DIG-labeled probes for in situ hybridization were constructed from RACE cDNA products of at least 500bp in p-GEM T Easy or from an existing EST library using the MEGAscript reverse transcriptase kit (AM1330 and AM1334) as previously described (Martindale et al., 2004).
Collection of embryos, DAPT treatment and In Situ Hybridization
N. vectensis adult polyps were spawned and embryos collected and de-jellied as previously described (Fritzenwanker and Technau, 2002). Embryos were raised in 1/3× filtered seawater (FSW) at 25C. Embryos in DAPT treatments were treated with 10μM DAPT (Sigma D5942) and 1% DMSO in 1/3× FSW for either 24 or 72 hours at 25C. Approximately 200 embryos per treatment were examined. Control embryos were treated with 1% DMSO in 1/3× FSW. Both control and DAPT-treated embryos were kept in the dark and the solution was changed every 12 hours (Kasbauer et al., 2007). Embryos were fixed in ice cold 4% paraformaldehyde and .2% gluteraldehyde in 1/3× FSW for 90 seconds followed by a second fixation of 4% paraformaldehyde in 1/3× FSW at 4C for 1 hour. Embryos were then rinsed 5 times in phosphate buffered saline with Tween-20 (PTw), one time in RNAse free dH20 and then washed 3× in MeOH and were stored in MeOH. In situ hybridization was conducted according to a previously published protocol (Martindale et al., 2004). Following in situ hybridization, embryos were mounted in 70% glycerol in 30% PBS and images were captured with a Zeiss AxioCam HRc camera.
cDNA construction and qPCR
RNA was extracted from wildtype, DAPT-treated, and morpholino injected embryos using the manufacturers recommended protocol with Trizol reagent (Invitrogen). Following RNA isolation, cDNA was constructed using 1.0μg of total RNA for each sample with Advantage RT for PCR kit (Clontech, Inc.). qPCR primers were designed for genes of interest to fragments of length 75-150bp (Supplemental File 1). The Nematostella gene GADPH was used as a control standard for all experiments. PCRs were conducted on an iCyclerCQ qPCR machine (BioRad).
SuH morpholino design and injection
A fluorescein-tagged morpholino antisense oligonucleotide (Gene Tools, Inc.) of sequence ATTACCAAAGCTGGACTGACCTTTT was designed to the splice donor site of the first intron of the su(H) transcript. Inclusion of the first intron results in normal translation of the first 34 residues of the wildtype protein as well as the insertion of four residues and a premature stop codon translated from intron sequence. The Su(H) morpholino was injected at a concentration of 0.5mM or 1.0mM using positive pressure injection. In order to assess the efficacy of the morpholino at blocking proper splicing, we designed a primer sequence flanking the first intron with a forward primer of sequence 5′ATGGACAGATATCTCACTGATGTAGTCC-3′ in the first exon and a reverse primer of sequence 5′-CATTTCTTGCTCACTGCTATTGCC-3′ in exon 2. Amplification of the wildtype transcript resulted in 249 bp fragment while amplification of a transcript in which the intron is included results in a 421 bp fragment.
Dominant Negative Generation and Injection
Messenger RNA coding for a dominant negative form of Suppressor of hairless, dnSu(H), was generated by using site-directed mutagenesis to replace codons specifying arginine and tyrosine residues of the DNA-binding domain of the SuH transcript (Supplemental Figure 2C) with codons specifying glutamic acid and serine respectively according to an approach employed in Xenopus (Wettstein et al., 1997). This construct was cloned into the pENTR dTOPO cloning vector (Invitrogen, Inc) and subsequently recombined into the destination vector consisting of a pRN3 backbone, carrying a c-terminal mCherry tag and Xenopus 5′ and 3′ hemoglobulin UTRs to form an expression vector (Roure et al., 2007). This expression vector was linearized and transcribed using Ambion mMESSAGE mMACHINE T3 kit. The RNA was subsequently purified with Ambion MEGAclear columns and injected into uncleaved embryos.
Cnidocyte, actin, and nuclear staining and imaging
Embryos used in cnidocyte staining experiments were fixed as above replacing 1/3× FSW with phosphate buffered saline (PBS) as previously described (Marlow et al., 2009; Szczepanek et al., 2002). Embryos used in phalloidin staining of f-actin and in nuclear staining were fixed as for in situ or as for cnidocyte staining with the exception that they were used within one week of fixation after storage at 4C in PBS and were not stored in MeOH. Embryos for cnidocyte staining were transferred to a solution containing DAPI and Tris buffer as previously described (Marlow et al., 2009; Szczepanek et al., 2002) which was modified to account for the molarity of 1/3× seawater (an additional 10g/L NaCl) and stained for 1 hour at room temperature. Embryos were then washed three times in PBS and stained in phalloidin and propidium iodide as previously described for cnidarian embryos (Magie et al., 2007; Marlow and Martindale, 2007). Embryos were viewed on a Zeiss Z-1 Axio-imager microscope and images were collected with an Orca-ER camera. Confocal imaging was conducted on a Zeiss LSM510 microscope running the LSM software package. Fluorescent images were false colored, cropped and γ corrected to visualize fainter staining internal structures using Volocity acquisition software (PerkinElmer Inc., Waltham, MA, USA) and Photoshop (Adobe Inc., San Jose, CA, USA).
Results
All components of the Notch signaling pathway are present in N. vectensis
N. vectensis Notch (NvNotch), like that of both flies and vertebrates, contains multiple EGF repeats that mediate the interaction with ligand molecules. A previously identified putative Notch homolog (Gazave et al., 2009) was further extended by EST data and conserved functional domains were predicted (Figure 1A and Supp. Fig. 1). While many EGF-containing genes are present in the sequenced genome, only a single gene with the diagnostic organization of well-conserved LNR domains, a NOD domain, transmembrane domain and Notch-like intracellular region was identified (Gazave et al., 2009). We were also able to identify a conserved nuclear localization signal in the C terminus of the protein (Figure 1A). The intracellular domain, which acts as a co-transcriptional activator when Notch is activated and cleaved, contains ankyrin repeats. While these domains can be found individually in many types of gene families throughout the Metazoa, it is the combination and relative order in addition to the characteristic LNR and NOD domains which identify the receptor specifically as Notch. Previous genome-wide surveys for Notch pathway members in N. vectensis have identified several putative delta-serrate ligand domain containing proteins (Gazave et al., 2009). By extending these putative gene models using RACE PCR and bioinformatic methods, we were able to confirm one of these genes to be the Delta ligand. NvDelta possessed all characteristic domains of bilaterian Delta or Serrate notch ligands: a DSL domain, EGF repeats and a transmembrane domain (Figure 1B). Several other DSL domain-containing ligands lacked EGF repeats which are necessary for mediating the known binding activities between the receptor Notch and Delta (Gordon et al., 2008). We were able to amplify two of these predicted ligands and extend their coding sequence by RACE PCR whereby it was possible to characterize their domain structures (Figure 1B). These DSL domain containing genes are referred to as Nv Repeated Delta Serrate Ligand (NvRdsl). We were able to revise the putative predicted transcript of NvRdsl2 (Gazave et al., 2009) and extend the predicted C terminal domain of NvRdsl1 (Figure 1B). Extension of NvRdsl1 revealed the presence of a transmembrane domain, indicating that the protein is likely to be membrane bound. This family of putative cell surface ligands is unique in that they contain multiple DSL domains and lack EGF domains and would therefore be predicted to serve as yet unknown roles.
Figure 1. All major components of the Notch signaling pathway are present inNematostella vectensis.
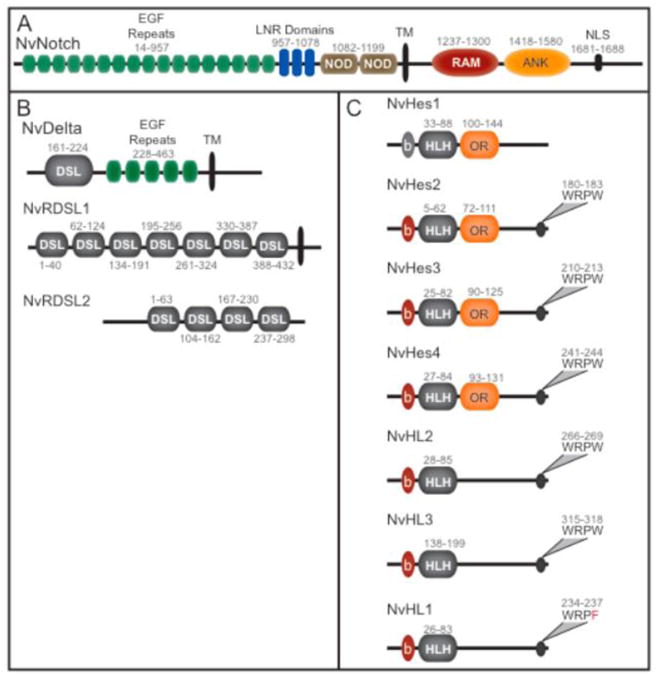
A, NvNotch is similar in organization and residue identity to those found in Bilaterian animals. B, Several DSL domain-containing sequences were isolated from N. vectensis. The Nematostella Delta contains one DSL domain, EGF repeats and a predicted transmembrane domain. Two additional DSL domain-containing sequences are unusual in the presence of multiple DSL domains. One of these sequences was found to contain a transmembrane domain. C, Four Hes genes (NvHes1-4) and three Hes-like (NvHl1-3) genes are present in N. vectensis. Conserved domains of the NvHes and NvHl genes are the bHLH domain (b and HLH), the orange domain (OR) and the C-terminal WRPW peptide. NvHes1 lacks a WRPW domain but maintains an orange and HLH domain. Positions of domains are indicated by numbers which refer to residues of the predicted protein.
Seven Hes basic-HLH containing transcription factors were identified based on conservation of the DNA binding domain (Figure 1C). Additional regions of the 5′ and 3′ coding regions were extended by RACE PCR. Three of these Hes genes contain both an orange domain that plays a role in mediating co-factor binding in addition to the bHLH domain (Kageyama et al., 2007) as well as a WRPW c-terminal domain necessary for Hes activity as a transcription repressor (Fisher et al., 1996), and could therefore be conclusively identified as Hes genes (Figure 1B). A fourth NvHes gene, NvHes1, contained an orange domain, a more divergent bHLH domain, but lacked the c-terminal WRPW domain. Other NvHes-like genes were also characterized. In NvHes-like 1(Nvhl1) the WRPW domain had been modified to WRPF (Figure IC). NvHes-like genes, NvHes-like 2(Nvhl2) and Nvhes-like 3(Nvhl3), were identified based on their bHLH domain and c-terminal WRPW domain characteristic of HES genes but both genes lacked the orange domain (Figure 1C).
A single locus for the Suppressor of Hairless transcription factor (NvSu(H)) is found in the N. vectensis genome. The corresponding predicted protein of this locus is extremely similar to that found in both protostome and deuterostome animals. The full-length coding region of NvSuH was verified by PCR from cDNA. All necessary DNA-binding domains are present in the Nematostella sequence and are required for the dual role of Su(H) as both a transcriptioncal activator and a transcriptional repressor (Supplemental Figure Figure 2A and 2B).
Notch pathway members are expressed throughout N. vectensis embryogenesis in overlapping domains
Transcripts encoding Nvnotch, Nvdelta and downstream effector molecules are first expressed as detected by in situ hybridization early in the gastrulation stage at approximately 20 hours post fertilization (hpf). The Nvnotch transcript is found broadly in both an oral and aboral ectodermal domain in the gastrula and early planula (Figure 2A and 2B) and later expands to include endodermal and pharyngeal endoderm and ectoderm expression in the later planula stage. Around the time of metamorphosis, the strongest Nvnotch expression is restricted to a narrow oral ring and to the primary larval neural structure, the apical tuft (Figure 2D).
Figure 2. Notch signaling components are expressed during gastrulation, planula and polyp formation inN. vectensis in overlapping domains in both ectodermal and endodermal tissues.
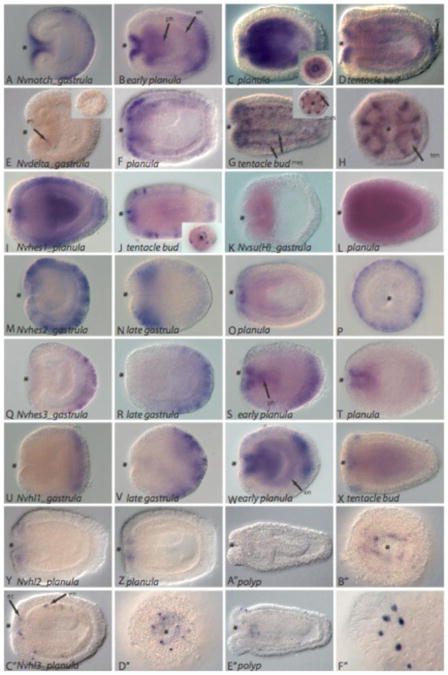
A, Notch is expressed at gastrulation and in the early planula (B) in oral (*) and aboral ectodermal domains. C, The expression domain in a later planula expands to include the developing pharynx and the endoderm. D, Late planula NvNotch expression is most pronounced in the ectodermal region surrounding the mouth and in the primary larval neural stucture, the apical tuft (at). Expression is also visible faintly throughout the endoderm. E, Nematostella Nvdelta is first expressed in a few cells in the blastula (inset) and early gastrula (arrow) and expands to include oral ectoderm and is also expressed in the endoderm (F). G, In the early polyp stage, expression is found in the developing mesenteries (mes) as pictured in a lateral view and oral view (inset). H, An oral view in which, Nvdelta is visible in the developing tentacle (ten) ectoderm. I-J, Nvhes1 is found expressed in aboral and lateral ectoderm and an oral ring of ectoderm as well as faintly within the endoderm of a planula. Within tentacle bud stage embryos, scattered ectodermal expressing cells are visible (I and J). J, (inset) An oral view of a tentacle bud stage embryo shows Nvhes1 expression around the oral opening (*) and surrounding ectoderm. K-L, NvSu(H) is expressed in the invaginating endodermal plate in an early stage gastrula (K) and faintly expressed throughout the ectoderm and endoderm of a planula stage (L). M, Nvhes2 is expressed throughout the developing embryonic ectoderm during gastrulation and is restricted to oral ectoderm in the late gastrula (N) and planula stage (O). P, An oral view of a gastrula stage embryo in which Nvhes2 is found in a large number of ectodermal cells. Q-X, Nvhes3 and Nvhl1 are initially expressed in developing aboral ectoderm during gastrulation and planula stages. During early planula stages (S and W), Nvhes3 and Nvhl1 expression domains are found in the aboral ectoderm and the pharynx and in the endoderm. T and X, In late planula stages, both Nvhes3 and Nvhl4 are expressed in scattered cells in the oral ectoderm and pharynx. Y-B″, Nvhl2 is expressed exclusively in a narrow expression domain surrounding the oral opening. C″-F″, Nvhl3 is expressed in scattered endoderm and ectodermal cells in planula (C″-D″) and polyp stages (E″-F″). D″ Oral view of Nvhl3 expressing cells in a planula (D″). F″, Ectoderm of a developing tentacle.
Nvdelta is expressed in a small number of scattered cells in the early gastrula stage (Figure 3E but is found widely expressed in the oral ectoderm and endoderm of the planula stage (Figure 3F). At metamorphosis, Nvdelta is restricted to tentacular ectoderm and the endodermal component of the developing mesenteries. Nvdelta encompasses a subset of the broader Nvnotch expression pattern domain (Figure 2C, 2D, 2F, 2G) and by the late planula and polyp stage is clearly expressed in distinct cells, while notch maintains low level expression in relatively large and diffuse territories. Transcripts for Nvsu(H), a requisite co-factor of the NICD transcriptional activator complex, are first expressed in the developing endodermal plate and later expressed ubiquitously throughout planula development (Figure 2K and 2L).
Figure 3. Treatment of embryos with the pharmacological agent DAPT leads to endodermal and pharyngeal defects and a loss of cnidocytes.
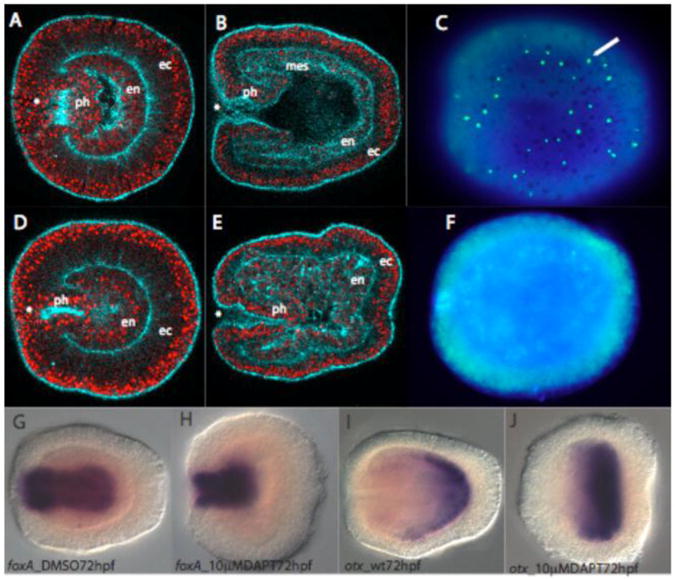
A-C, Control embryos treated with 1% DMSO at 24 (A) and 78 hpf (B and C). F-actin is marked with phalloidin (blue) and nuclear staining is visible propidium iodide (red). C, A surface view in which cnidocyte cells (green) are visible throughout the ectoderm which is counterstained with DAPI nuclear stain (blue). D-F, Embryos treated with 10μM DAPT in which 24 hpf embryos complete gastrulation (D) but fail to form normal larval structures by 78 hpf (E). F, Cnidocyte cells are absent in a surface view of a 78hpf embryo. G and H, Nvforkhead is expressed in the developing pharynx of both DMSO control (G) and DAPT treated (H) embryos. I and J, Nvotx, a control gene normally expressed in aboral endoderm, is expressed in both DMSO controls (I) and DAPT treated (J) embryos.
As would be predicted for effectors of Notch signaling, the sum of all Nvhes and Nvhes-like transcripts largely recapitulate the expression of the notch receptor. Nvhes1 is strongly expressed in ectodermal cells in the planula and polyp stages (Figure 2I-2J), and is also expressed at low levels in endodermal cells in the planula (Figure 2I). Nvhes2 is expressed in a large number of cells throughout the ectoderm of the gastrula stage (Figure 2M). In planula stages, Nvhes2 is restricted to the oral ectodermal domain. Nvhes3 (Figure 2Q-2T) and Nvhl1 (Figure 2U-2X) are expressed exclusively in aboral ectoderm of the gastrula and early planula but expand to include both oral ectoderm and endoderm in the planula stage and are restricted orally by the late planula stage. Nvhes3 and Nvhl1 form a syn-expression group, while Nvhl2 is restricted to only the oral domain of this expression territory and NvHes1 becomes restricted to single cells within this territory. Nvhes2 is largely distinct from the expression of Nvhes3, Nvhl1 and Nvhl2, particularly with respect to the large oral ectoderm domain of expression. By the late planula and tentacle bud stage and the overlay of Nvhes2, Nvhes3 and Nvhl1 appear to encompass the entire notch expression pattern. In contrast, Nvhes1 (Figure 2I-2L), Nvhl2 (Figure 2Y-2B″) and Nvhl3 (Figure 2C″-2F″) expression extend later into development and are characterized by staining in a smaller number of very strongly expressing cells. Interestingly, Nvhl2 and Nvhl3 lack an orange domain, while the DNA-binding region of Nvhes1 is lacking the characteristic proline in the basic region indicating that they may utilize novel co-factors for DNA-binding or may have acquired new transcriptional targets.
Notch signaling and the Su(H) transcription factor are required for morphogenesis and cell fate
In order to determine the role of active Notch signaling in N. vectensis development we utilized the pharmacological agent and inhibitor of γ-secretase, DAPT (Dovey et al., 2001) which prevents proteolytic cleavage of the Notch receptor thereby blocking the internalization and subsequent activity of the NICD as a transcriptional activator (Geling et al., 2002). Approximately 100 embryos per concentration were treated with 1, 5, 10, and 20μM DAPT to determine appropriate DAPT concentrations for subsequent experiments. Those embryos treated with 10μM DAPT showed consistent morphological phenotypes and high viability, while those at higher concentrations showed signs of toxicity, therefore all subsequent experiments were conducted with the 10μM concentration of DAPT.
N. vectensis embryos were treated with 10μM DAPT or a control DMSO treatment directly following fertilization and were assayed at 24, 48, 72, 96 and 144hpf. By 24hpf both control and DAPT treated embryos had successfully undergone gastrulation (Figure 3A and 3D). By 72hpf discernable phenotypic deficiencies emerge in DAPT treated embryos wherein growth of the pharynx is stunted, mesentery formation does not occur, and endodermal morphology is disrupted when compared to control embryos (Figure 3B and 3E). While the endoderm of control embryos is a monolayer (Figure 3B), the endoderm of DAPT-treated embryos is stratified and disorganized (Figure 3E). Ectodermal cell fates are also altered with cnidocyte stinging cells completely absent in DAPT treated embryos (Figure 3C and 3F). As embryos appear to successfully undergo gastrulation in both treatments we used markers for the boundary between ectoderm and endoderm, the winged helix transcription factor NvfoxA (Figure 3G and 3H), and the aboral endoderm marker Nvotx which confirm that endodermal patterning is intact in embryos of both treatments at 72hpf (Figure 4I-4J). Nvotx expression begins early in development and marks a large aboral endodermal region while NvfoxA expression marks the developing pharynx and the ectodermal margin of the ecodermal-endodermal boundary (Magie et al., 2007). This indicates that the initial regional patterning following gastrulation occurs and is maintained but that later development and morphogenesis are affected.
Figure 4. Injection of a morpholino directed against the transcription factor Su(H) and a dominant negative Su(H) result in similar morphological phenotypes to those observed in DAPT-treated embryos.
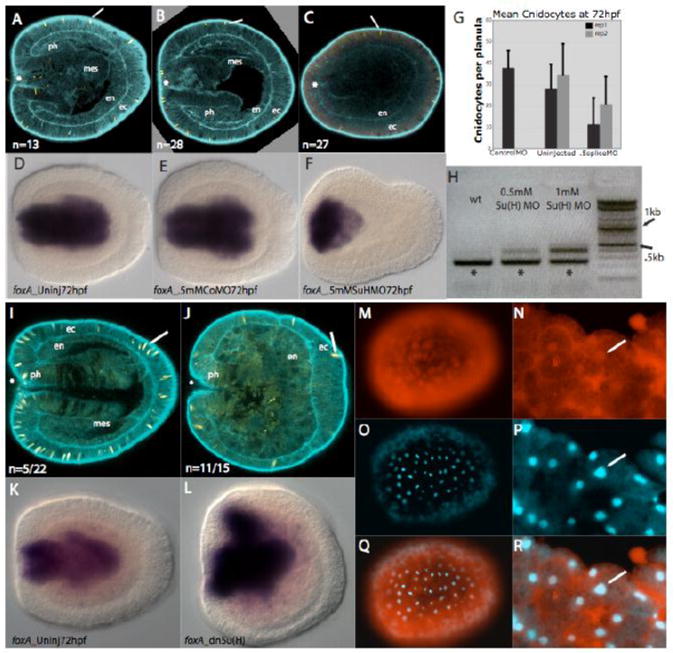
A, An uninjected wildtype 72 hpf embryo. B, Embryo injected with 0.5mM control morpholino which is indistinguishable from wildtype (A). C, An embryo injected with 0.5mM splice-blocking Su(H) morpholino in which the pharynx is absent, mesenteries are absent and a greatly reduced number of cnidocyte cells are visible. D-F, forkhead, control gene, expression in the pharynx of uninjected, control morpholino-injected and NvSu(H) morpholino-injected embryos. In all three treatments, forkhead is strongly expressed in the developing pharynx. In the case of F, where the pharynx does not differentiate, NvfoxA is expressed in the specified pharyngeal tissue. G, A graph showing the mean number of cnidocytes in uninjected embryos and those injected with control or NvSu(H) splice-blocking morpholino in two different experiments. Embryos injected with NvSu(H) morpholino have significantly fewer cnidocytes than those which remained uninjected or those that were injected with control morpholino. Twelve embryos for each treatment in each trial were examined. H, Amplification of cDNA constructed from wildtype (wt) embryos and those injected with 0.5mM and 1.0mM NvSu(H) splice-blocking morpholino with primers in exon1 and exon2. A band made up of wildtype transcript (*) and a larger band representing transcript in which the first intron is included are visible in the morpholino injected samples. A faint band representing trace amounts of pre-mRNA is visible in uninjected controls. I and J, F-actin is marked with phalloidin (blue) and cnidocytes are stained with DAPI (yellow). Oral is to the left (*), and endoderm (en), ectoderm (ec), mesenteries (mes) and pharynx (ph) are indicated. I, 72hpf control embryo in which larval structures including cnidocytes, pharynx and mesenteries are visible . J, A 72hpf embryo injected with dominant negative 500ng/uL NvSu(H) in which the pharynx is greatly reduced, mesenteries are absent and cnidocyte number is reduced. K and L, In situ hybridization to the control gene forkhead in which the developing pharynx is stained in both 72hpf control (K) and 72hpf dnSuH injected (L) embryos. M-R, Live blastula stage N. vectensis embryos in which the dnSuH protein and mCherry tag are visibly expressed (red) and nuclear staining is visible (DAPI). M, Blastula stage embryo with tagged dnSuH protein visible. N, High magnification image of blastula stage embryo with visible dnSuH protein in the cytoplasm and nucleus (arrow). O, Blastula stage embryo with visible nuclear staining in same embryo as above (M). P, High magnification of blastula stage nuclear staining in same embryo as above (N). Q and R, (Q) is an overaly of M and O, while (R) is an overlay of N and P. Arrow indicates a cell nucleus. Numbers in panels A-C indicate the total number of embryos stained and examined for general morphology. Fractions in I and J indicate the number of embryos displaying an abnormal disorganized endoderm out of the total number of injected embryos examined.
In current models of canonical Notch signaling, once cleaved, the NICD directly interacts with the Su(H) transcription factor to effect transcription of downstream targets. Knockdown of expression levels of the Su(H) transcription factor should therefore produce similar phenotypes to those produced through chemical inhibition of Notch receptor cleavage with DAPT for those phenotypes linked to canonical Notch signaling. To reduce levels of the NvSu(H) transcription factor we employed a morpholino antisense oligonucleotide to block splicing of the Nvsu(H) transcript and introduce a premature stop codon following the first coding exon. rtPCR analysis of Nvsu(H) transcripts in control and injected embryos confirms a dose-responsive effect on transcript splicing (Figure 4H). The 1mM and 0.5mM Su(H) morpholino concentrations give rise to the largest proportion of unspliced transcript, but some toxicity was detected in the control morpholino injected embryos at 1mM concentration, therefore lower concentrations were used for subsequent experiments. Embryos injected just after fertilization were assayed at 72hpf and found to have a poorly developed pharyngeal region and to lack mesenteries (Figure 4A-4C), and to have significantly fewer cnidocytes (Figure 4G). Normal expression of the control gene NvfoxA was observed however, indicating that pharyngeal tissue was regionally specified but did not properly differentiate (Figure 4D-4F).
We also knocked-down Su(H) function by overexpressing a dominant negative Nvsu(H) transcript. This transcript codes for a mutated DNA-binding site which renders the dnNvSu(H) protein unable to bind the upstream regions of target genes (Supplemental Figure 2A). The dominant negative transcript dnNvsu(H) was fused to a 3′ sequence encoding the fluorescent C-terminal mCherry tag and the expressed protein showed nuclear expression in injected embryos (Figure 4M-4R). Morphological phenotypes of embryos injected with dominant negative su(H) transcript (Figure 4I and 4J), like those resulting from Su(H) morpholino injection, showed disorganized endoderm, failure of mesentery formation, and a reduced number of cnidocytes. Expression of the NvfoxA marker indicates proper acquisition of pharyngeal tissue identity following gastrulation (Figure 4K and 4L).
The splice-blocking NvSu(H)-targeted morpholino was effective in producing a partial block of splicing in embryos but did not result in a complete knockdown of wildtype transcript (Figure 4H) and would not be effective against any maternally deposited protein or maternally deposited transcript. In addition, the dnSu(H) construct was only stably expressed, as assayed by the fluorescence of the mCherry tag, for the first one to two days of development and was therefore not sufficient to produce detectable fluorescence indicative of expression of the protein later in development. For these reasons, we chose to use chemical inhibition of Notch signaling in subsequent experiments to assess the role of Notch signaling on cell-type specification and boundary formation.
Hes gene expression is regulated by Notch signaling in Nematostella
In both vertebrate and protostome taxa, the Hes gene family of transcriptional repressors are the primary effectors of Notch signaling events making them obvious candidates when examining targets of N. vectensis Notch signaling. In situ hybridization of 24hpf and 72hpf DAPT and DMSO treated embryos show significantly reduced hes gene expression (Figure 5A-5J and Supplemental Figure 3) at both time points as well as changes in spatial gene expression when compared to wildtype expression (Figure 5B, 5D, 5F, 5H, 5J and 5K). NvHes1 expression is found only in the oral ectoderm of 72hpf DAPT treated embryos (Figure 5D), an expression domain which is normally turned off following the gastrula stage of wildtype embryos (Figure 5K). NvHl1 expression is broadly and faintly expressed in oral ectoderm of 72hpf DAPT treated embryos (Figure 5H), and is not restricted to the more narrowly expressed wildtype pattern (Figure 5G). Quantitative PCR analysis of cDNA constructed from DAPT treated versus DMSO control-treated embryos reveals a knockdown in hes transcript levels at 72hpf (Figure 5L). Proneural genes act as effectors of Notch-mediated control of neurogenesis through their position as downstream targets of Hes genes. In the presence of Notch signaling, one described role of the Hes genes is to act as repressors of proneural gene expression (Kageyama et al., 2008). We assayed four proneural-like genes from Nematostella (Simionato et al., 2007) following DAPT treatment and found these proneural-like genes to be upregulated (Figure 5L).
Figure 5. Nvhes and hes-likegene expression is reduced when Notch signaling is disrupted with DAPT.
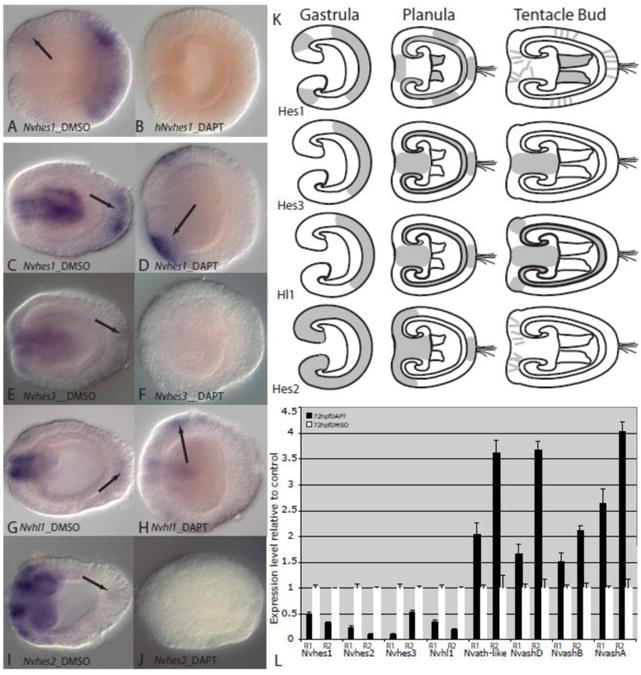
A-D, Nvhes1 expression is present in aboral ectoderm and faintly in oral ectoderm (arrow) in 24hpf 1% DMSO control embryos (A) but absent in 24hpf 10μM DAPT treated embryos (B). Similar losses of expression in DAPT treated embryos are seen for Nvhes3 and Nvhl1 (Supplementary Figure 3). C, Nvhes1 expression is present in pharyngeal ectoderm and aboral ectoderm of 1% DMSO treated 72hpf embryos but reduced and expressed (arrow) in oral ectoderm in 10μM DAPT treated embryos (D). E-F, Nvhes3 expression is detectable in pharyngeal tissues in 72hpf controls (F) but expression is faintly detected in pharyngeal tissues and oral ectoderm (arrow). G-H, Nvhl1 is detectable in both control (G) and DAPT treated 72hpf embryos (H) but is greatly reduced and is found in oral ectoderm (arrow) in DAPT treated embryos. I, Nvhes2 is expressed in oral and pharyngeal ectoderm in control embryos, but is absent in 72hpf DAPT treated embryos (J). K, Summary of wildtype expression patterns of Nvhes1-3 and Nvhl1 in gastrula, planula and tetacle bud stages. L, qPCR results for Nvhes and Nvhl genes and proneural-like genes in DAPT treated embryos and control embryos for two replicates (R1 and R2). Changes in expression level are indicated by fold expression in relation to control DMSO-treated embryos which were assigned an expression level of one. Standard error bars were calculated using three replicate reactions for each sample.
Disruption of Notch cleavage results in a “neurogenic” phenotype
Recently, a number of markers of neural development have been identified in Nematostella vectensis (Anctil, 2009; Marlow et al., 2009). We used these markers to assay the role of Notch signaling in specifying the neural cell fate in N. vectensis embryos (Figure 6). RNA-binding proteins msi and elav are expressed in elements of the developing nervous system. In wildtype embryos, elav is found predominantly in the developing endodermal nervous system at 72hpf while msi is found in oral ectodermal cells. Both msi (Figure 6A-6D) and elav (Figure 6E-6H) are expressed in a much larger population of cells in 72hpf DAPT-treated embryos than in control embryos. In 72hpf DAPT-treated embryos, elav expression is primarily ectodermal which is typically indicative of much earlier stages in wild type embryos in which elav shifts from early ectodermal to later stage endodermal staining. In addition, elav is expanded to include a large region of the apical pole ectoderm in the region of the apical tuft and the ectodermal region of the pharynx (Figure 6G). gcm, which is expressed primarily in endodermal cells of control 72hpf embryos (Figure 6I and 6J), is widely ectopically expressed in aboral ectodermal cells of DAPT treated embryos (Figure 6K and 6L). paxA (Figure 6M-6P) and paxC (Figure 6Q-6T), molecular markers of ectodermal sensory neurons, are also broadly expanded at 72hpf. paxC is expressed in a small number of oral ectodermal cells in control embryos (Figure 6Q and 6R). In DAPT-treated embryos, scattered oral ectodermal staining is present in addition to an ectopic aboral ectodermal domain (Figure 6S and 6T). While expression of all neural markers assayed was dramatically expanded in 72hpf DAPT-treated embryos, no increase of neural marker expression could be detected in 24hpf DAPT-treated embryos (Figure 6Y-6B″), suggesting that the MO loses its effectiveness over time.
Figure 6. Disruption of Notch signaling with DAPT results in an upregulation of neural marker genes.
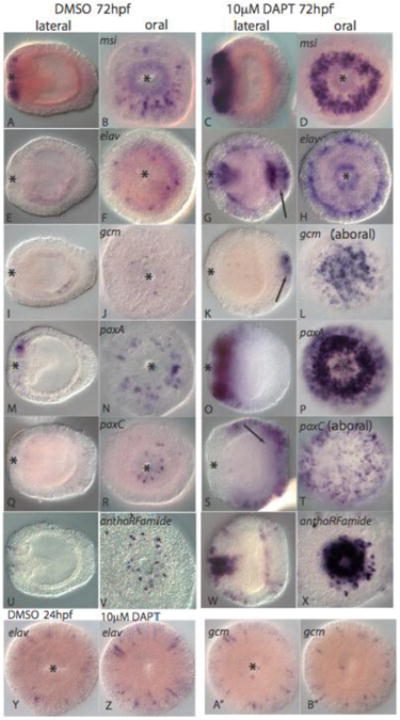
Panels on the left are control embryos in lateral (left) and oral (second from left) views treated with DMSO while those on the right are treated with DAPT except for L and T which are aboral views. A-D, msi is expressed in oral ectodermal cells in both control (A and B) and DAPT treated embryos (C and D) but is expressed in many more cells in DAPT treated embryos. E-H, A small number of endodermal cells express the elav transcript in control DMSO treated embryos (E and F) while a much larger number of cells in both ectodermal and endodermal tissues of DAPT treated embryos express elav (G and H). I-L, In control embryos gcm is expressed in scattered endodermal cells (I) and a small number of oral ectodermal cells (J) in control embryos but is expressed in a large number of aboral ectodermal cells in DAPT treated embryos (K and L). M-P, paxA expression expands in oral ectoderm cells in DAPT treated embryos (O and P) in comparison to control embryos (M and N). Q-T, paxC is expressed in a small number of oral ectodermal cells in control embryos (Q and R) but is expressed in a large number of aboral ectodermal cells DAPT treated embryos (S and T). U-X, Neurons expressing the peptide antho-RFamide in control embryos (U and V) and DAPT treated embryos (W and X). Y-B″, In 24hpf embryos, the number of cells expressing elav and gcm is the same in DMSO (U and V) and DAPT treated (W and X) embryos.
We also examined the number of differentiated neurons expressing the anthozoan neuropeptide NvanthoRFamide in embryos in which Notch cleavage was blocked with DAPT and those which were injected with the Nvsu(H) splice-blocking morpholino. In both 24hpf and 72hpf DAPT-treated embryos there was an increase in the number of NvanthoRFamide expressing neurons (Figure 6U-6X), however this increase was much more dramatic in 72hpf embryos (Figure 6Y-6X). In contrast to DAPT-treated embryos at 72hpf, we did not observe an increase in NvanthoRFamide expressing neurons in embryos injected with the Su(H) morpholino (Supplemental Figure 4).
Notch signaling has a late role in partitioning the developing tentacle field
In order to assess the late function of Notch signaling in morphogenesis, we treated embryos with the γ-secretase inhibitor DAPT from 3 to 5 days post-fertilization in the period directly preceding and overlapping with the early settlement phase. During settlement, the body column elongates and tentacles grow from the prospective tentacle buds of ectoderm surrounding the mouth. We find that embryos treated with DAPT fail to partition the tentacle ectoderm into the four tentacle fields, as marked by the tentacle ectoderm marker paxA (Figure 7G-7K). In some animals, the ectodermal tentacle field, as marked by the expression of paxA, remains joined by a semicircle of expression (Figure 7J) and in others a complete ectodermal NvpaxA positive ring remains (Figure 7K). Consistent with these expression studies, we find that some animals, which have been allowed to recover and develop to the polyp stage, show two fused tentacles (Figur 7C and 7D) while more severe cases are found to have all four tentacles fused (Figure 7E). Animals injected with lower doses of morpholino, which have been raised to the polyp stage, also display this phenotype with varying numbers of fused tentacles (Figure 7B).
Figure 7. Inhibition of Notch signaling results in defects in tentacle formation.
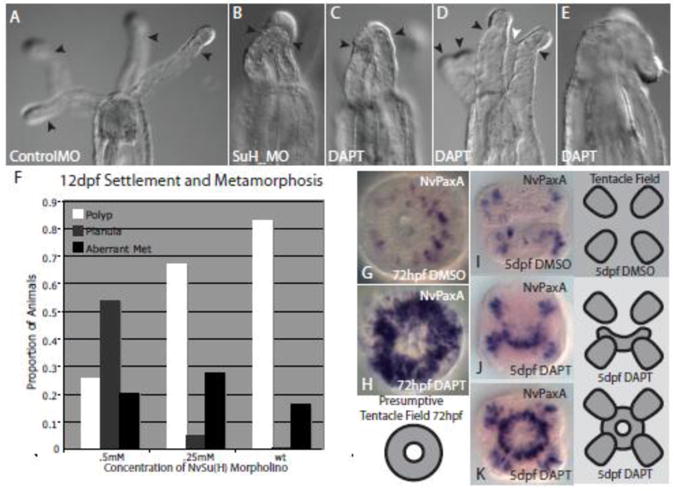
Polyps at 12dpf. A, Animals injected with control morpholino show normal tentacle formation as the oral tentacle field has split into four individual tentacles (black arrows). B, Polyps injected with the Su(H) morpholino display two or more tentacles which remain unsplit resulting in a fused tentacle phenotype. Embryos which were treated with DAPT during tentacle formation and then recovered to late polyp stages also show similar fused tentacle morphologies with two (C and D) or four fused tentacles (E). D, In some individuals, a thin epithelia can be seen connecting two fused tentacles (white arrow). F, The number of animals which metamorphosed to fully formed polyps in relation to those which failed to settle or displayed abnormal morphologies in control and morpholino injected animals. G, The tentacle field as demarcated with the oral ectodermal marker paxA in wildtype 72hpf embryos versus those treated with DAPT (H). I, A control embryo showing the tentacle field marked with paxA. Polyps which were treated from 3 to 5 days with DAPT showed partial (J) or complete (K) failure of the tentacle field to split. Illustrations depict the developing tentacle field before and during tentacle bud formation. In DAPT-treated animals, this field fails to fully split and tentacles remain fused.
Discussion
Canonical Notch signaling controls mesentery formation, cnidogenesis, and boundary formation in N. vectensis
notch expression is broad in N. vectensis embryos and encompasses several tissues including the pharyngeal (ectodermal and endodermal components), body wall endodermal (gastrodermal plus mesenterial tissue) and ectodermal regions (Figure 2A-2D) as can be expected from its many functions in other taxa. In contrast, expression of Nvdelta (Figure 2E-2H) and Nvhes and Nvhl transcripts (Figure 2I-2F″) are restricted to region-specific expression patterns. Interestingly, Nvdelta in the ectoderm, is restricted to the staining of scattered cells reminiscent of the developing spirocyte cells (Marlow et al., 2009). Upon planula settlement, the Delta ligand is expressed nearly exclusively in the mesenteries (body wall endoderm) and ectodermal portion of the tentacles that gives rise to spirocytes (Figure 2G and 2H). Oral ectoderm of the planula and the body wall endoderm that forms the mesenteries of the developing polyp are both encompassed by notch expression, and the scattered Nvdelta-expressing cells lie within the Nvnotch expressing tissues. Mesenteries harbor the cnidoglandular tracts which give rise to developing nematocysts and the tentacle tips house the developing spirocyst stinging cells (Marlow et al., 2009; Williams, 1979). Several different stages of developing cnidocytes can be found in the cnidoglandular tracts, indicating active cell differentiation from a resident stem population. Although less is known about the development of spirocyst cnidocytes in the tentacles, a similar scenario of local differentiation from a progenitor population is likely considering the absence of any data to support long-range cell migrations in N. vectensis.
Functional data support a Notch-mediated and SuH-mediated role in cnidogenesis. Blocking Notch signaling through inhibition of γ-secretase results in a complete loss of cnidocytes. Morpholino mediated knock-down and dominant negative inhibition of Su(H), an effector of Notch signaling, similarly result in a substantial loss of cnidocytes. Involvement of both the Notch receptor and the SuH transcription factor in a loss of cnidocyte phenotype suggest that the canonical Notch signaling pathway, implicated in many Notch-mediated developmental events, plays an important role in cnidogenesis. In C. elegans, Drosophila and vertebrates, canonical Notch signaling targets the Ref-1, E(spl), and Hes families respectively (Borggrefe and Oswald, 2009; Iso et al., 2003; Neves and Priess, 2005). These genes are all bHLH proteins which are evolutionarily descended from an ancestral Hes-like gene family and have a conserved role in Notch signaling across the Bilateria (Simionato et al., 2007). Our examination of the expression of six of these Nvhes and Nvhl genes in Nematostella revealed the transcription of at least four genes Nvhes1, Nvhes2, Nvhes3, and Nvhl1 are likely to be downstream of canonical Notch signaling. Further studies will need to be conducted to determine which of these Hes genes, or combination thereof will be responsible for the cnidocyte phenotype in N. vectensis. Blocking Notch cleavage through antagonism of γ-secretase produces a much stronger Hes gene knockdown than that produced through MO knockdown of Su(H). We attribute this discrepancy either to the presence of maternal transcript or an incomplete block of splicing by the morpholino (Figure 5H). Notch mediates partitioning of the tentacle field in Nematostella and may indicate a conserved role for Notch signaling in the demarcation of morphological territories. Notch has a role in segment formation in both vertebrates and arthropods and has been recently shown to mediate bud separation in asexual budding of the cnidarian hydra (Munder et al., 2010). This indicates that in both cnidarians and bilaterians, and therefore what is inferred to be the case for the eumetazoan ancestor, partitioning of morphological territories was among the ancestral roles of the pathway. These conserved developmental roles of the Notch pathway in Nematostella, as has been demonstrated in Hydra (Kasbauer et al., 2007; Munder et al., 2010), include this boundary formation as well as a role in cnidogenesis. As the neurogenic Notch effects observed in Nematostella have not been demonstrated in the cnidarian Hydra, it is likely that Notch signaling is more divergent between anthozoan cnidarians and hydrozoan cnidarians. Given the widespread utilization of Notch signaling in metazoan neurogenesis, it is likely that the neurogenic Notch signaling role was lost from Hydra but maintained in anthozoa.
Neurogenic phenotypes occur after blocking Notch signaling
Broad expression of the Notch receptor in ectodermal tissues suggests additional roles for Notch signaling outside of the cnidogenic pathway. We assayed several classes of putative neural markers, all of which were heavily upregulated in 72 hpf embryos in which Notch signaling had been blocked by DAPT. These neural markers showed particularly large increases in the number of cells expressing the marker in the oral territory and in the apical tuft (Figure 6). Presence of additional NvanthoRFamide positive cells confirms that the number of differentiated neurons present in N. vectensis also increases substantially. Neurogenic phenotypes are nearly universal in Bilaterians in which Notch signaling has been disrupted. Usually, these phenotypes are mediated by Su(H) directed transcriptional regulation. In N. vectensis, a DAPT-mediated block of all Notch signaling results in a substantial increase in the number of cells expressing neural markers. Su(H) MO knockdowns produce a detectable neurogenic phenotype only in the cnidocyte stinging cells and produce defects in normal gastrodermal morphogenesis but not other aspects of neurogenesis as assayed with these markers (Fig. 6). Notch has been previously reported to act independently of SuH mediated pathways, which might explain the different phenotypes observed between Notch and Su(H) knockdowns (Hori et al., 2004; Sanalkumar et al.). However, it is also likely that remaining wildtype transcript may account for the differences between DAPT and morpholino phenotypes in Nematostella. In addition, DAPT may interfere with the activity of other developmentally important pathways which could also account for differences between the DAPT chemical treatments and Su(H)-targeted experiments.
In contrast to the marked increase in neural cell types we observe in Nematostella, no alterations to neurogenesis were observed in the adult cnidarian Hydra after DAPT treatment (Kasbauer et al., 2007). There are several likely explanations for the differences observed in Nematstella and Hydra DAPT-induced phenotypes. Importantly, experiments conducted in Hydra were performed in adult animals, not developing embryos, and are likely to reflect the condition of Notch signaling in adult stem cell populations. Secondly, Hydra neurogenesis and that observed in anthozoans is quite distinct as endodermally derived i-cell populations and cell migrations are not observed in anthozoans (Marlow et al., 2009). Finally, it is noteworthy that a genome-wide study of the Nematostella and Hydra genome reveal loss and substantial divergence of Notch pathway components in Hydra in comparison to other metazoan taxa, including Nematostella (Gazave et al., 2009). Molecular conservation of the N. vectensis Notch pathway as well as the accessibility to embryonic development strongly promotes N. vectensis as a cnidarian model for future Notch signaling studies.
There has been significant elaboration of the Notch pathway in Nematostella
Both non-metazoan taxa (such as choanoflagellates) as well as all previously examined metazoans contain proteins representing the core components of the Notch membrane-bound receptor. In metazoans, these domains became uniquely organized into the characteristic Notch architecture (King et al., 2008). In addition, the first Notch ligands, Delta and Serrate can be found throughout the metazoa, but are not found in non-metazoan eukaryotes (Gazave et al., 2009). A single proneural gene and a hes-like gene are also present in sponges with Notch, delta and this proneural gene all appearing to have neurogenic function when injecting in vertebrate embryos (Richards et al., 2008). As sponges do not appear to have definitive neurons, it is unclear what their role in sponge development might be.
While the core components of the Notch signaling pathway in N. vectensis occur as single copy genes, there has been the generation of additional target genes and potential ligands. Single Nvnotch , Nvjagged/serrate and Nvdelta loci occur in N. vectensis (Gazave et al., 2009) but we confirmed the presence of multiple Nvhl and Nvhes genes and two additional DSL-domain bearing putative Notch ligands predicted in the sequenced genome by rtPCR and in situ hybridization. Nematostella hes genes have undergone duplication to give rise to the four hes genes and three hl genes (characterized here). While several of these genes are found in tandem repeats in the genome, further evidence suggests that they may represent recent duplications (Gazave et al., 2009) that have diverged in both expression and sequence and are likely to play different roles in development. One of the newly described DSL ligands is predicted to be membrane bound while the other was not found to contain a recognizable transmembrane region (Figure 1B). Diffusibile Notch ligands are not described in vertebrates or Drosophila but were recently described in the nematode C. elegans (Chen and Greenwald, 2004; Komatsu et al., 2008). As in Nematostella, C. elegans possesses both diffusible and membrane bound proteins. Little is currently known about their function, but they are hypothesized to interact with previously described ligands of the C. elegans Notch pathway. A similar scenario is possible in Nematostella and may provide additional modes of regulations for Notch activation in different developmental contexts.
Duplications of members of gene families implicated in neural development as well as terminal differentiation markers of neurons are emerging as common features of the N. vectensis genome. Such anthozoan-specific duplications have occurred in the Pax gene family (Matus et al., 2007a), the proneural gene family of bHLH transcription factors (Simionato et al., 2007), the opsin family of molecules in cnidarian photoreceptors (Marlow, unpublished) and the shaker (Kv1) potassium channels (Jegla and Marlow, unpublished). It is currently unclear what role duplication may serve in cnidarian neural evolution but it is likely that the lack of centralized neural integration could contribute to the development of functionally specialized independent neural circuits.
Supplementary Material
Highlights.
Spatial characterization of Notch pathway components in anthozoan N. vectensis
Functional evidence for ancient role of Notch in neurogenesis and cnidogenesis
bHLH transcription factors downstream of Notch in cnidarian Notch signaling pathway
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anctil M. Chemical transmission in the sea anemone Nematostella vectensis: A genomic perspective. Comparative Biochemistry and Physiology-Part D: Genomics and Proteomics. 2009;4:268–289. doi: 10.1016/j.cbd.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Ang HL, Tergaonkar V. Notch and NFkappaB signaling pathways: Do they collaborate in normal vertebrate brain development and function? Bioessays. 2007;29:1039–1047. doi: 10.1002/bies.20647. [DOI] [PubMed] [Google Scholar]
- Arias AM. New alleles of Notch draw a blueprint for multifunctionality. Trends Genet. 2002;18:168–170. doi: 10.1016/s0168-9525(01)02635-x. [DOI] [PubMed] [Google Scholar]
- Barolo S, Walker RG, Polyanovsky AD, Freschi G, Keil T, Posakony JW. A notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Greenwald I. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev Cell. 2004;6:183–192. doi: 10.1016/s1534-5807(04)00021-8. [DOI] [PubMed] [Google Scholar]
- Chipman AD, Akam M. The segmentation cascade in the centipede Strigamia maritima: involvement of the Notch pathway and pair-rule gene homologues. Dev Biol. 2008;319:160–169. doi: 10.1016/j.ydbio.2008.02.038. [DOI] [PubMed] [Google Scholar]
- Darling JA, Reitzel AR, Burton PM, Mazza ME, Ryan JF, Sullivan JC, Finnerty JR. Rising starlet: the starlet sea anemone, Nematostella vectensis. Bioessays. 2005;27:211–221. doi: 10.1002/bies.20181. [DOI] [PubMed] [Google Scholar]
- Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Latimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, Mutter LC, Nietz J, Quinn KP, Sacchi KL, Seubert PA, Shopp GM, Thorsett ED, Tung JS, Wu J, Yang S, Yin CT, Schenk DB, May PC, Altstiel LD, Bender MH, Boggs LN, Britton TC, Clemens JC, Czilli DL, Dieckman-McGinty DK, Droste JJ, Fuson KS, Gitter BD, Hyslop PA, Johnstone EM, Li WY, Little SP, Mabry TE, Miller FD, Audia JE. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- Finnerty JR, Pang K, Burton P, Paulson D, Martindale MQ. Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science. 2004;304:1335–1337. doi: 10.1126/science.1091946. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Molecular and cellular biology. 1996;16:2670. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzenwanker JH, Technau U. Induction of gametogenesis in the basal cnidarian Nematostella vectensis(Anthozoa) Dev Genes Evol. 2002;212:99–103. doi: 10.1007/s00427-002-0214-7. [DOI] [PubMed] [Google Scholar]
- Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic-Licina M, Chera S. Origins of neurogenesis, a cnidarian view. Dev Biol. 2009;332:2–24. doi: 10.1016/j.ydbio.2009.05.563. [DOI] [PubMed] [Google Scholar]
- Gazave E, Lapebie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, Borchiellini C, Vervoort M, Renard E. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol Biol. 2009;9:249. doi: 10.1186/1471-2148-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A -secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO reports. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Arnett KL, Blacklow SC. The molecular logic of Notch signalingña structural and biochemical perspective. Journal of Cell Science. 2008;121:3109. doi: 10.1242/jcs.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- Hori K, Fostier M, Ito M, Fuwa TJ, Go MJ, Okano H, Baron M, Matsuno K. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Development, growth & differentiation. 2008;50:S97–S103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- Kasbauer T, Towb P, Alexandrova O, David CN, Dall'armi E, Staudigl A, Stiening B, Bottger A. The Notch signaling pathway in the cnidarian Hydra. Dev Biol. 2007;303:376–390. doi: 10.1016/j.ydbio.2006.11.022. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelzer S, Klein T. A Notch-independent function of Suppressor of Hairless during the development of the bristle sensory organ precursor cell of Drosophila. Development. 2003;130:1973–1988. doi: 10.1242/dev.00426. [DOI] [PubMed] [Google Scholar]
- Komatsu H, Chao MY, Larkins-Ford J, Corkins ME, Somers GA, Tucey T, Dionne HM, White JQ, Wani K, Boxem M, Hart AC. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 2008;6:e196. doi: 10.1371/journal.pbio.0060196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- Lee PN, Kumburegama S, Marlow HQ, Martindale MQ, Wikramanayake AH. Asymmetric developmental potential along the animal-vegetal axis in the anthozoan cnidarian, Nematostella vectensis, is mediated by Disheveled. 2007 doi: 10.1016/j.ydbio.2007.05.040. [DOI] [PubMed] [Google Scholar]
- Lee PN, Pang K, Matus DQ, Martindale MQ. A WNT of things to come: evolution of Wnt signaling and polarity in cnidarians. Semin Cell Dev Biol. 2006;17:157–167. doi: 10.1016/j.semcdb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Lewis J, Hanisch A, Holder M. Notch signaling, the segmentation clock, and the patterning of vertebrate somites. J Biol. 2009;8:44. doi: 10.1186/jbiol145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Magie CR, Daly M, Martindale MQ. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Dev Biol. 2007;305:483–497. doi: 10.1016/j.ydbio.2007.02.044. [DOI] [PubMed] [Google Scholar]
- Marlow HQ, Martindale MQ. Embryonic development in two species of scleractinian coral embryos: Symbiodinium localization and mode of gastrulation. Evol Dev. 2007;9:355–367. doi: 10.1111/j.1525-142X.2007.00173.x. [DOI] [PubMed] [Google Scholar]
- Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, Martindale MQ. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev Neurobiol. 2009;69:235–254. doi: 10.1002/dneu.20698. [DOI] [PubMed] [Google Scholar]
- Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa) Development. 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Go MJ, Sun X, Eastman DS, Artavanis-Tsakonas S. Suppressor of Hairless-independent events in Notch signaling imply novel pathway elements. Development. 1997;124:4265–4273. doi: 10.1242/dev.124.21.4265. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Pang K, Daly M, Martindale MQ. Expression of Pax gene family members in the anthozoan cnidarian, Nematostella vectensis. Evol Dev. 2007a;9:25–38. doi: 10.1111/j.1525-142X.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Thomsen GH, Martindale MQ. FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev Genes Evol. 2007b;217:137–148. doi: 10.1007/s00427-006-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickey KM, Mello CC, Montgomery MK, Fire A, Priess JR. An inductive interaction in 4-cell stage C. elegans embryos involves APX-1 expression in the signalling cell. Development. 1996;122:1791–1798. doi: 10.1242/dev.122.6.1791. [DOI] [PubMed] [Google Scholar]
- Munder S, Kasbauer T, Prexl A, Aufschnaiter R, Zhang X, Towb P, Bottger A. Notch signalling defines critical boundary during budding in Hydra. Dev Biol. 2010;344:331–345. doi: 10.1016/j.ydbio.2010.05.517. [DOI] [PubMed] [Google Scholar]
- Neves A, Priess JR. The REF-1 family of bHLH transcription factors pattern C. elegans embryos through Notch-dependent and Notch-independent pathways. Dev Cell. 2005;8:867–879. doi: 10.1016/j.devcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Pourquie O. The vertebrate segmentation clock: the tip of the iceberg. Curr Opin Genet Dev. 2008;18:317–323. doi: 10.1016/j.gde.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Schultz J, Milpetz F, Bork P. SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res. 1999;27:229–232. doi: 10.1093/nar/27.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U. FGF signaling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development. 2008;135:1761–1769. doi: 10.1242/dev.020784. [DOI] [PubMed] [Google Scholar]
- Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Current Biology. 2008;18:1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- Roure A, Rothb‰cher U, Robin F, Kalmar E, Ferone G, Lamy C, Missero C, Mueller F, Lemaire P. A multicassette Gateway vector set for high throughput and comparative analyses in ciona and vertebrate embryos. PLoS ONE. 2007;2:e916. doi: 10.1371/journal.pone.0000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanalkumar R, Dhanesh SB, James J. Non-canonical activation of Notch signaling/target genes in vertebrates. Cell Mol Life Sci. 67:2957–2968. doi: 10.1007/s00018-010-0391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F. Suppressor of Hairless is required for signal reception during lateral inhibition in the Drosophila pupal notum. Development. 1995;121:1875–1884. doi: 10.1242/dev.121.6.1875. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, McClay DR. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development. 1999;126:1703–1713. doi: 10.1242/dev.126.8.1703. [DOI] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P. Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development. 1990;109:509–519. doi: 10.1242/dev.109.3.509. [DOI] [PubMed] [Google Scholar]
- Szczepanek S, Cikala M, David CN. Poly-gamma-glutamate synthesis during formation of nematocyst capsules in Hydra. J Cell Sci. 2002;115:745–751. doi: 10.1242/jcs.115.4.745. [DOI] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, Bince JM, Xu R, Martindale MQ. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–450. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- Williams RB. Studies on the nematosomes of Nematostella vectensis Stephenson (Coelenterata: Actiniaria) Journal of Natural History. 1979;13:69–80. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


