Fig. 4. Conformational changes of the sub-viral particle that trigger transcription.
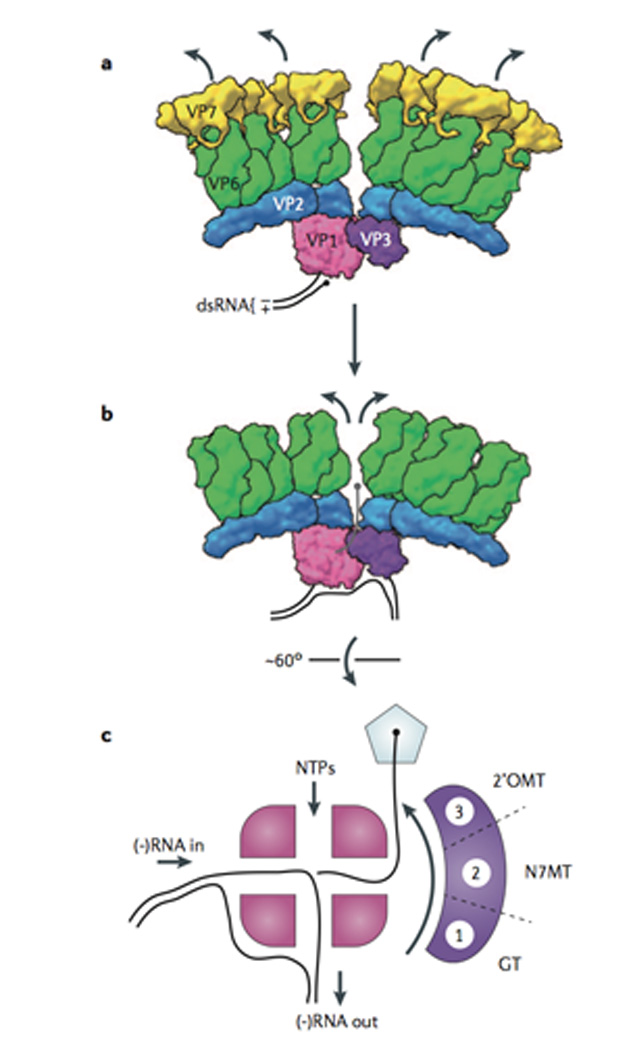
(A) Model for the transcriptionally inactive rotavirus virion. Prior to entry, the VP7 layer (yellow) appears to suppress transcription by the viral polymerase complex (PC), VP1 (pink) and VP3 (purple)3, 4. Current data suggest that a single PC resides near almost all of the 5-fold channels in a rotavirus particle16. In this inactive state, the RNA-dependent RNA polymerase VP1 (PDB ID 2R7U) is likely bound to the 3’ end of the (-) strand of the dsRNA, poised for transcription of the (+)RNA10. (B) The VP7 layer (PDB ID 3IYU) dissociates during entry, resulting in the upward- and outward-movement of the VP6 (green) and VP2 (blue) layers (PDB ID 3N09) of the particle3, 54. This movement results in expansion of the 5-fold channel and permits extrusion of (+)RNA transcripts4. (C) Model for the passage of the nascent (+)RNA transcript through the PC and out of the particle. The four tunnels of VP1 permit the input (-)RNA and NTPs to support synthesis of the new (+)RNA, and the re-formation of the parental dsRNA pair10. The 5’ end of the (+)RNA must rapidly be recruited and capped (black sphere) by VP3 prior to exit through the 5-fold channel (pentagon). Biochemical data, and comparison with the BTV capping enzyme (VP4; used here as a surrogate for rotavirus VP3; PDB ID 2JH8)50, suggest that VP3 is a multi-domain protein that successively modifies the 5’ end through (1) guanylyltransferase, (2) N-7-methyltransferase, and (3) 2’-O-methyltransferase activities to generate the mature rotavirus 5’ cap structure.
