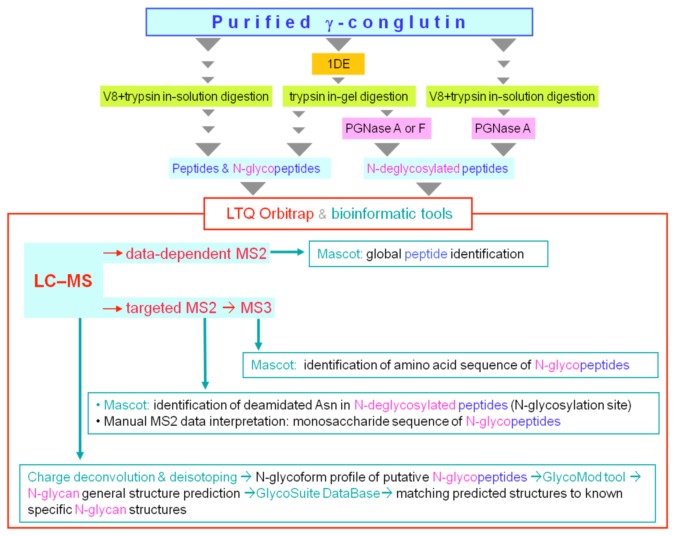
Figure 1. Schematic overview of the glycoproteomic workflow.
Combinations of the following procedures were used: 1) reducing SDS-PAGE to isolate the N-glycosylated large subunit, 2) γ-conglutin proteolytic digestion in-gel (with trypsin) or in-solution (with endopeptidase GluC (V8) followed by trypsin) to generate different mixtures of peptides and N-glycopeptides; 3) N-deglycosylation of the digests with PNGase A or PNGase F to identify the N-glycosylation site, and assess the presence/absence of “core” α1-3 fucose. The digests (with or without N-deglycosylation) were then analyzed by LC–Orbitrap MS, with MS survey scans followed by data-dependent ITMS2 or targeted MSn. Mass spectral data analysis included the use of: 1) automated charge-deconvolution of high resolution-high mass accuracy spectra, and isotope envelope simulation; 2) bioinformatic tools for in silico glycoform structure prediction (GlycoMod and GlycoSuiteDB), and database search (Mascot) for sequence identification of non-glycosylated or enzymatically deglycosylated peptides; (3) manual inspection of MS2 and MS3 spectra of glycopeptides for sequence annotation of their monosaccharide and amino acid components, respectively.