Figure 5. sX13 activity does not require Hfq.
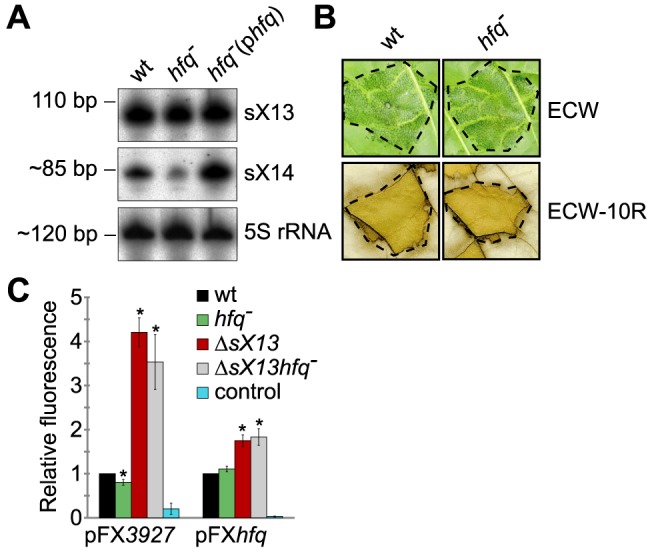
(A) Northern blot analysis. Total RNA from NYG-grown Xcv strains 85-10 (wt), the hfq frameshift mutant (hfq−) and the hfq mutant ectopically expressing Hfq (phfq) was analyzed using sX13- or sX14-specific probes. 5S rRNA was probed as loading control. The experiment was performed twice with two independent mutants and with similar results. (B) Plant infection assay. The Xcv wild-type 85-10 (wt) and hfq mutant strain (hfq−) were inoculated at 2×108 cfu/ml into leaves of susceptible ECW and resistant ECW-10R plants. Disease symptoms were photographed 6 dpi. The HR was visualized 2 dpi by ethanol bleaching of the leaves. Dashed lines indicate the inoculated areas. The experiment was repeated three times with similar results. (C) GFP fluorescence of NYG-grown Xcv 85-10 (wt), the hfq mutant (hfq−), the sX13 deletion mutant (ΔsX13) and the double mutant (ΔsX13hfq−) carrying pFX3927 or pFXhfq. Xcv autofluorescence was determined by Xcv 85-10 carrying pFX0 (control). Data points and error bars represent mean values and standard deviations obtained with at least four independent experiments. GFP fluorescence of the wt was set to 1. Asterisks denote statistically significant differences (t-test; P<0.01).
