Figure 6. sX13 loops impact on Xcv virulence.
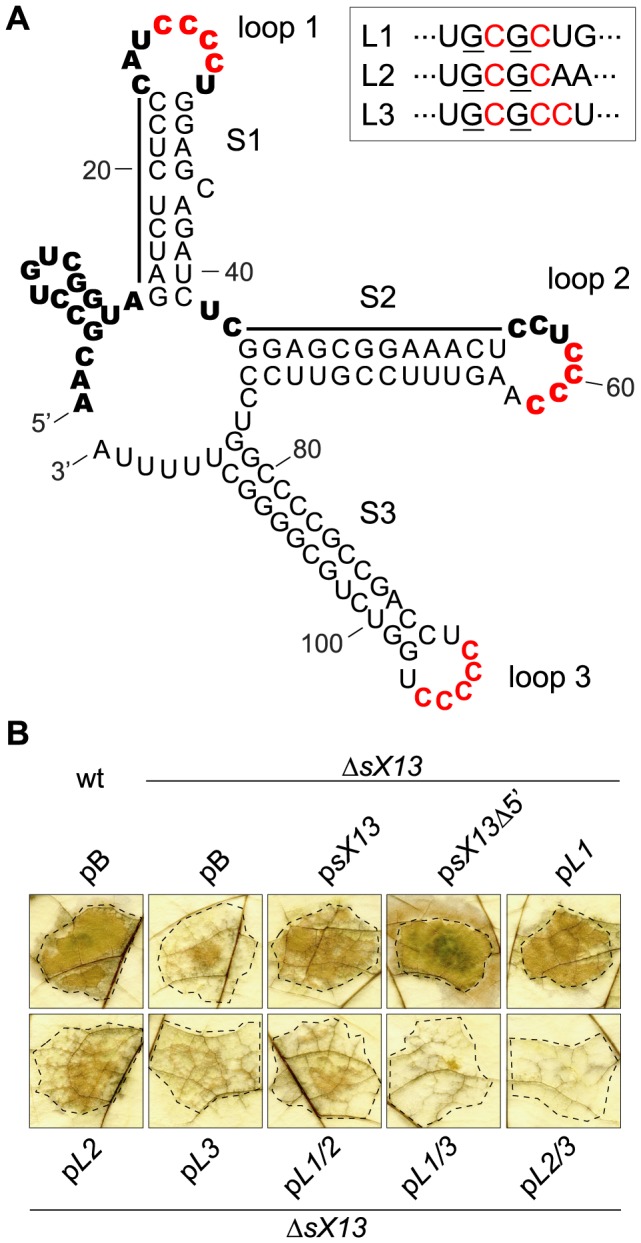
(A) Secondary structure of sX13 based on prediction and probing (see Figure S2). sX13 consists of an unstructured 5′-, three double-stranded regions (S1; S2; S3) and three loops (loop 1–3). 4C-/5C-motifs are highlighted in red. Bold letters indicate unpaired bases and bars mark double-stranded regions deduced from structure probing. Mutations in loops are boxed, exchanged nucleotides are underlined. (B) Derivatives mutated in loops 2 and 3 fail to complement the plant phenotype of ΔsX13. Leaves of resistant ECW-10R plants were inoculated at 108 cfu/ml with Xcv 85-10 (wt) and ΔsX13 carrying pBRS (pB), psX13 or one of the following derivatives: sX13 lacking 14 terminal nucleotides (psX13Δ5′), sX13 mutated in single loops (pL1, pL2, pL3) or in two loops (pL1/2, pL1/3, pL2/3). The HR was visualized by ethanol bleaching of the leaves 2 dpi. Dashed lines indicate the inoculated areas. The experiment was performed four times with similar results.
