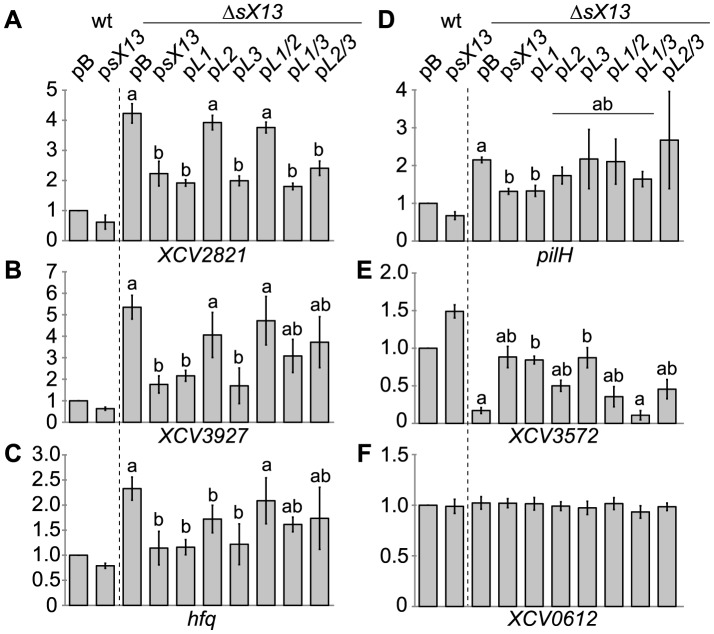
Figure 7. sX13 loops differentially contribute to abundance of putative mRNA targets.
Relative transcript levels of (A) XCV2821, (B) XCV3927, (C) hfq, (D) pilH, (E) XCV3572 and (F) XCV0612 were analyzed by qRT-PCR in total RNA of NYG-grown Xcv strains 85-10 (wt) and ΔsX13 carrying pBRS (pB), psX13 or mutated sX13-derivatives (see Figure 6). The mRNA abundance in the wt was set to 1. Data points and error bars represent mean values and standard deviations obtained with at least three independent biological samples. Statistically significant differences are indicated (t-test; P<0.015).