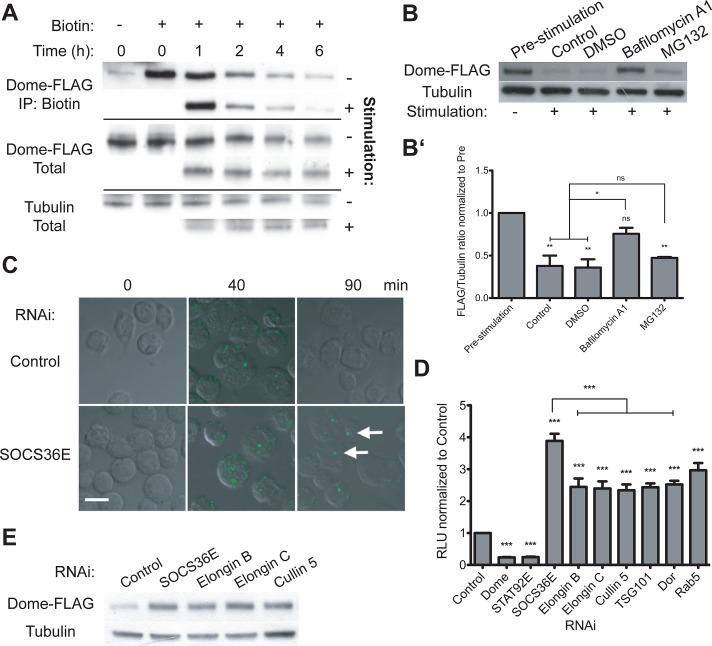
FIGURE 1:
Dome stability and the role of Elongin B/C, Cullin-5, and SOCS36E. (A) Levels of total and biotinylated Dome-FLAG present in cyclohexamide-treated cells following stimulation with either mock or Upd2-GFP–conditioned medium at the indicated time points. (B and B′) Levels of Dome-FLAG following a 2-h stimulation, as indicated. By comparison with untreated and DMSO carrier controls, pre-incubation with 0.2 μM bafilomycin A1 prevented ligand-induced degradation of Dome, while treatment with 10 μM MG132 had no significant effect. (B′) The average (and SEM; **, p < 0.001; *, p < 0.05) of three experiments, with (B) showing a representative example. (C) Drosophila Kc167 cells treated with the indicated dsRNAs prior to stimulation with Upd2-GFP (green). After allowing for ligand internalization, low-pH washes removed remaining plasma-membrane-bound ligand prior to incubation for the indicated times. By contrast with controls, cells lacking SOCS36E still include ligand structures (white arrows), even 90 min after stimulation. (D) JAK/STAT pathway activity as shown by the 6×2xDrafLuc reporter is modulated by dsRNAs targeting the indicated genes. Knockdown of Elongin B/C, Cullin-5, and proteins required for multiple stages of endocytic trafficking all results in broadly equivalent increases in pathway activity. Dome and STAT92E knockdowns act as controls. Significance is shown for selected combinations with ***, p < 0.0001; **, p < 0.001; *, p < 0.05. (E) By comparison with control cells, knockdown of SOCS36E, Elongin B/C, and Cullin-5 stabilize Dome under steady-state conditions.