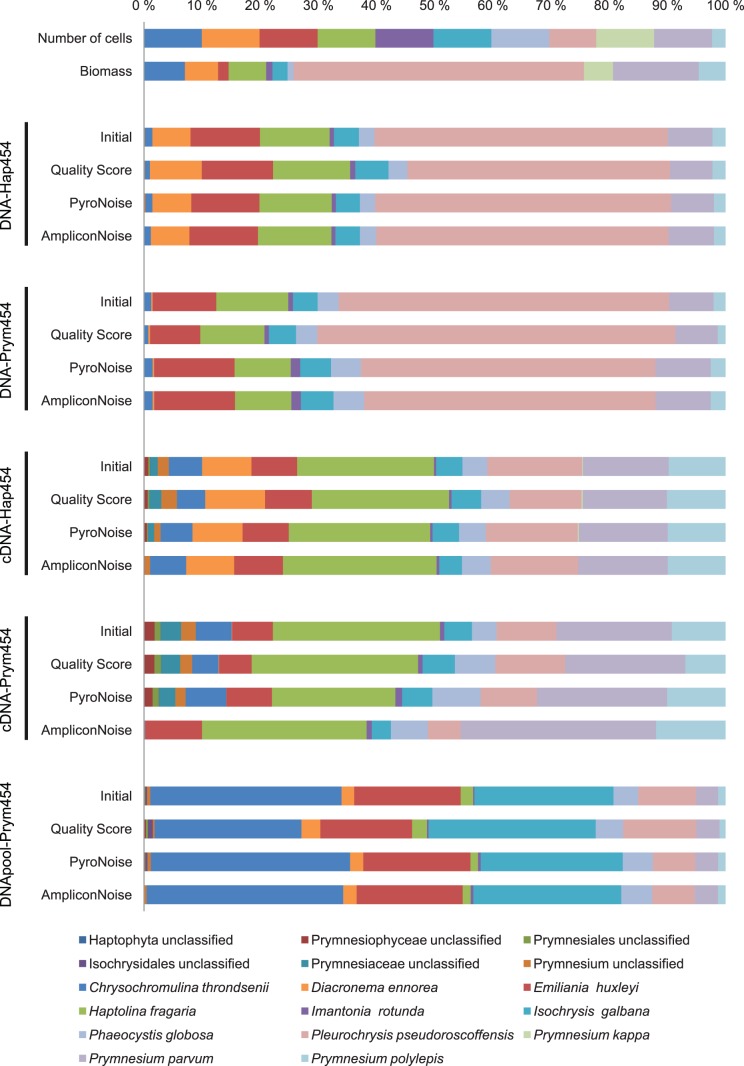
Figure 1. Proportional species abundance in 454 pyrosequencing reads from a mock community of haptophytes.
Compared to the initial distribution of species in terms of cell number (first row) and biomass (second row) in the mock community. The different combinations of template (DNA or RNA/cDNA), primer pair, (Hap454 or Prym454), and read filtering strategy are indicated on the figure. Initial Filtering (IF): Initial filtering based on match with barcode and primer, and removal of reads with Ns and/or homopolymers >8 bp, and reads that are shorter than 365 bp. Quality Score (QS): requiring minimum average quality score 30 over a 50 bp moving window along the read, in addition to IF. PyroNoise (PN): the PyroNoise algorithm as implemented in mothur, with default settings. AmpliconNoise (AN): AmpliconNoise as implemented in Qiime v.1.4.0, with default settings. Chimera identification by Perseus was applied to all datasets. The filtering strategies are described in the methods section.