Figure 1. The founding of C. rubella and the identification of its founding haplotypes.
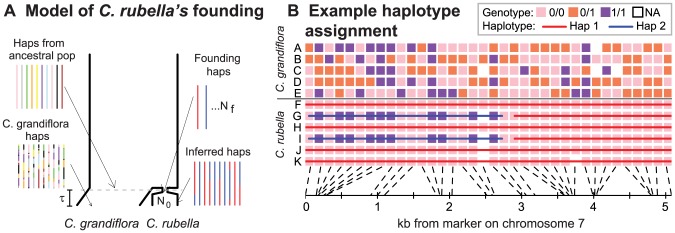
A) A cartoon coalescent model of C. rubella's origin. At time,  , a population ancestral to C. rubella is formed by sampling
, a population ancestral to C. rubella is formed by sampling 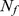 chromosomes (i.e. haplotypes, haps) from a large outcrossing population ancestral to both species, and this selfing population quickly recovers to size,
chromosomes (i.e. haplotypes, haps) from a large outcrossing population ancestral to both species, and this selfing population quickly recovers to size, 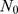 . Because some of the
. Because some of the 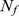 lineages are lost to drift, we can identify the founding haplotypes surviving to the present, which we color in red and blue. While recombination scrambles ancestral chromosomes in C. grandiflora, the low effective recombination rate in C. rubella ensures that large chunks of founding haplotypes remain intact. B) We aim to identify these founding haplotypes by using patterns of sequence variation (see text and METHODS for details of our algorithm). Here, we present an example of founding haplotype identification in a typical genomic region. To aid visualization, we label the major allele in C. rubella as ‘0’, and the allele that is rare or absent C. rubella as ‘1’, and only display genotypes at sites with common variants in C. grandiflora. In the left hand side of Figure 1B, there are clearly two distinct founding haplotypes on the basis of patterns of variation at sites polymorphic in both species. On the right hand side, all C. rubella individuals are identical at sites polymorphic in C. grandiflora, so we infer a single founding haplotype.
lineages are lost to drift, we can identify the founding haplotypes surviving to the present, which we color in red and blue. While recombination scrambles ancestral chromosomes in C. grandiflora, the low effective recombination rate in C. rubella ensures that large chunks of founding haplotypes remain intact. B) We aim to identify these founding haplotypes by using patterns of sequence variation (see text and METHODS for details of our algorithm). Here, we present an example of founding haplotype identification in a typical genomic region. To aid visualization, we label the major allele in C. rubella as ‘0’, and the allele that is rare or absent C. rubella as ‘1’, and only display genotypes at sites with common variants in C. grandiflora. In the left hand side of Figure 1B, there are clearly two distinct founding haplotypes on the basis of patterns of variation at sites polymorphic in both species. On the right hand side, all C. rubella individuals are identical at sites polymorphic in C. grandiflora, so we infer a single founding haplotype.
