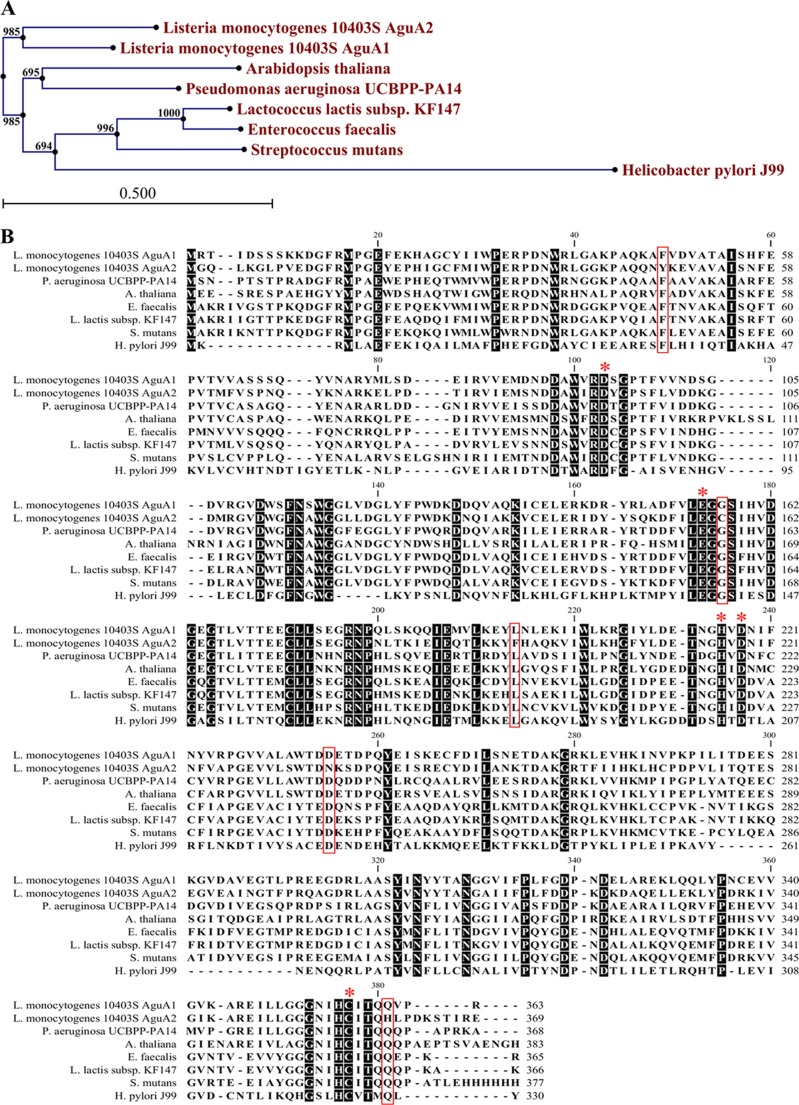
FIGURE 2.
Bioinformatic analysis of AguA1 and AguA2. A, phylogenetic tree of AguA1/AguA2 and homologs from other bacterial species. The tree was constructed by the neighbor-joining program, and a bootstrap test of 100 replicates was used to estimate the confidence of branching patterns, where the numbers on internal nodes are the support values. B, amino acid sequence alignment of AguA1/AguA2 against homologs from A. thaliana, H. pylori, S. mutans, E. faecalis, L. lactis, and P. aeruginosa. The conserved motifs and residues are shaded. The key amino acid residues noted with asterisks are involved in substrate binding and enzyme activity. The boxed residues are different between AguA2 and AguA1 or other AgDIs.