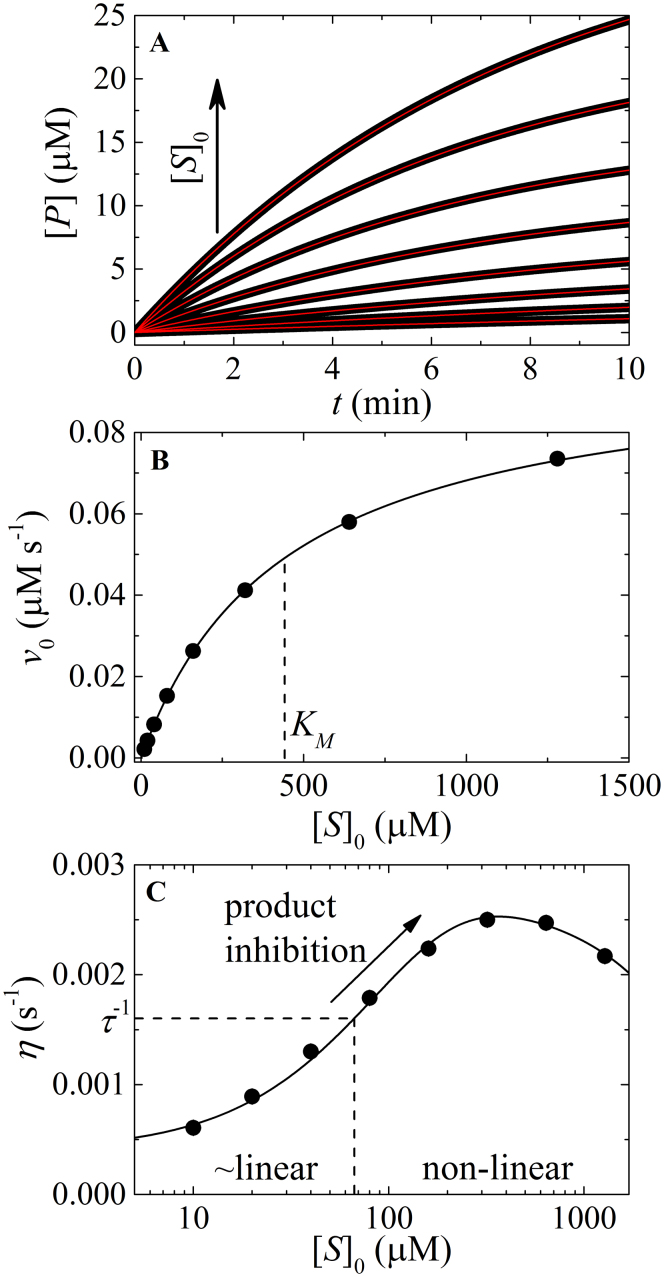
Figure 2. Analysis of non-linear enzyme kinetic time courses.
(A). Simulated enzyme cycling time courses according to Figure A1 with initial [substrate] (from bottom to top) of 10, 20, 40, 80, 160, 320, 640, and 1280 μM, and the following fundamental reaction rate constants: k+1 = 0.06 μM s−1, k−1 = 25 s−1, k+2 = 0.1 s−1, k−2 = 0.3 s−1, k+3 = 10 s−1, k−3 = 1 μM s−1, k+4 = 10 s−1, k−4 = 0.1 μM s−1, k+5 = 10 s−1, k−5 = 0.1 μM s−1, k+6 = 9.7 s−1, and k−6 = 0.8 μM s−1. The solid lines through the simulated data represent the best-fits to the product inhibition/substrate depletion equation (Eq. 1). (B). [S]0-dependence of the initial enzyme cycling velocity. The solid line represents the best-fit to a rectangular hyperbola (Eq. A9), yielding values of Vm = 0.098 (±0.0005) μM s−1, KM = 442.8 (±4) μM, which agree well to the values of Vm = 0.097 μM s−1, KM = 412.2 μM predicted from the fundamental rate constants (Eq. A10). (C). [S]0-dependent enzyme cycling velocity reduction rate constant η. The solid line through the data points is for visualization only. Substrate concentration regimes where time courses will display linear and non-linear behavior are indicated.