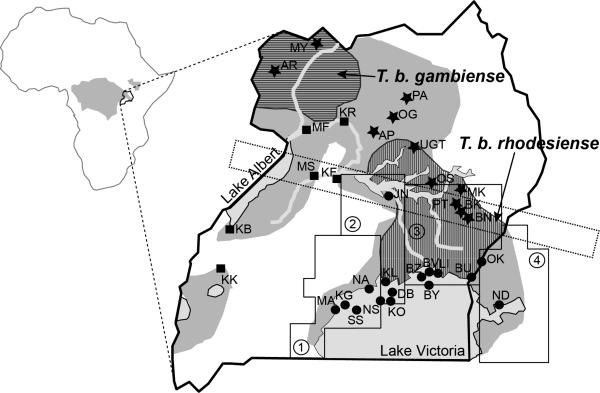
Figure 1. Glossina fuscipes and HAT distribution in Uganda.
Left: Map of Africa showing in gray the G. fuscipes distribution and the location of Uganda. Right: Map of Uganda with Gff sampling sites that are referred to in the text. Different shades of gray identify main water bodies (light gray) and the current Gff distribution (gray). The ranges of the human infective Trypanosoma brucei, T. b. rhodesiense and T. b. gambiense, are superimposed on the Gff distribution and identified with a vertical and horizontal hatched pattern, respectively. The populations analyzed genetically in previous studies are identified with two letters and a symbol for their assignment to one of the three main genetic clusters (dot=southern; star=northern; square= western). The dashed rectangle identifies the contact zone around Lake Kyoga in central Uganda where the northern and southern mtDNA haplogroups co-occur. The four operational blocks identified by PATTEC to progressively eradicate Gff from the Lake Victoria basin from west to east are also identified with small dashed lines and numbers [90,91]. The blocks are based on the Food and Agriculture Organization (FAO) predicted habitat suitability for Gff, natural barriers, major urban areas, international borders, and drainage patterns. Block 1 in the western part of the country is the most isolated block due to the expansion of the city of Kampala and subsequent urbanization and habitat fragmentation of the surrounding area. Block 2 in the central area has been targeted for control to create a buffer zone between the eradication block and the rest of the Gff predicted range in Uganda, while only vector population monitoring activities are planned for the other two blocks during phase 1 of the control program. Upon successful eradication in block 1, block 2 would become the eradication target and so on until the whole basin is tsetse-free.