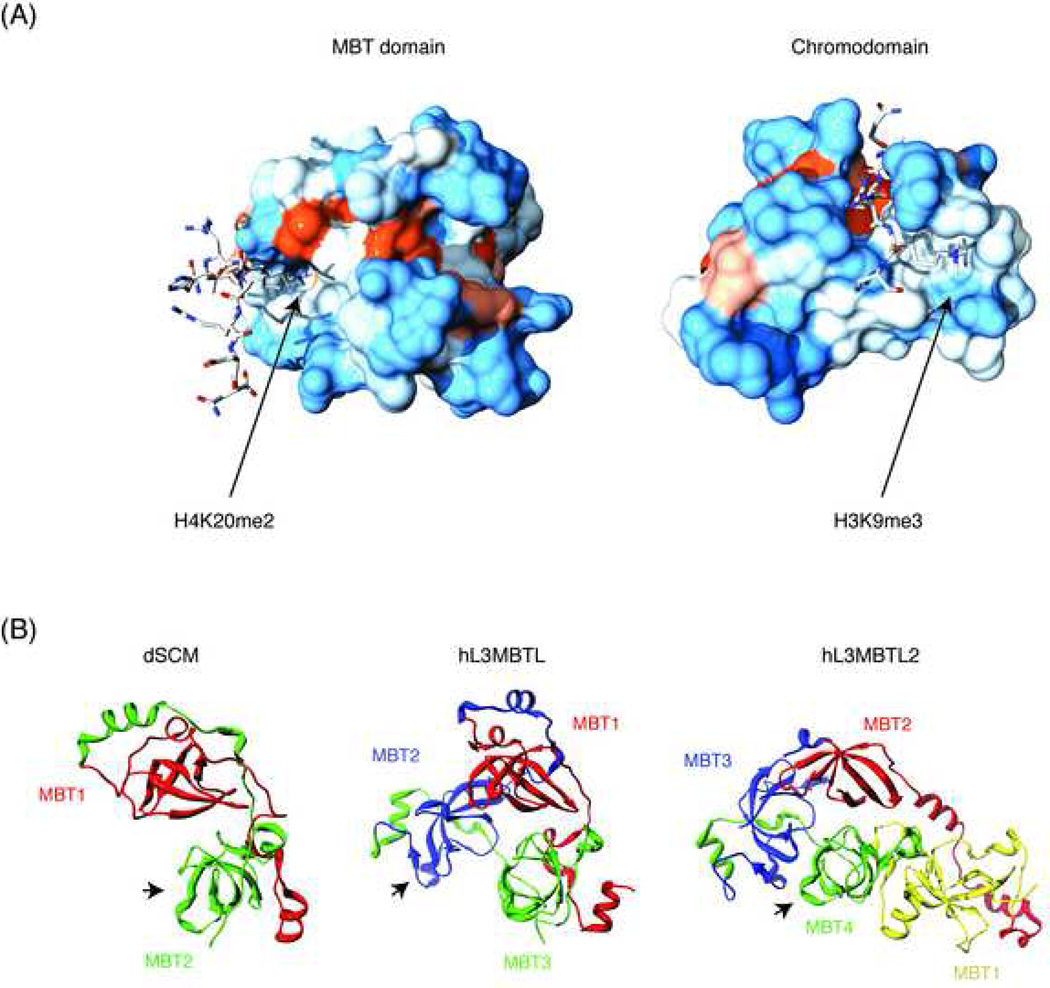
Figure 2. Structural features of the MBT repeats.
(A) The crystal structure of the 2nd MBT repeat of hL3MBTL is shown in complex with the ligand H4K20me2 (left) and compared with a similar structure obtained for the chromodomain of dHP1 in complex with H3K9me3 (right). The two different modes of binding, pocket insertion versus surface groove are illustrated. Figures were generated with UCSF chimera and POV raytracing software using atomic coordinates from PDB entries 2PQW and 1Q3L. Surface colors indicate hydrophobicity according to the Kyte-Doolittle scale, ranging from blue for the most hydrophilic to red for the most hydrophobic. (B) Schematic depiction of higher order organization of 2, 3, and 4 tandem MBT repeats, as found in SCM, L(3)MBT and SFMBT family proteins. Structurally homologous domains across families are in the same color. Known binding sites for histone peptides are marked with an arrow. The atomic coordinates used were from PDB entries 2R5A, 1OZ2 and 3F70.