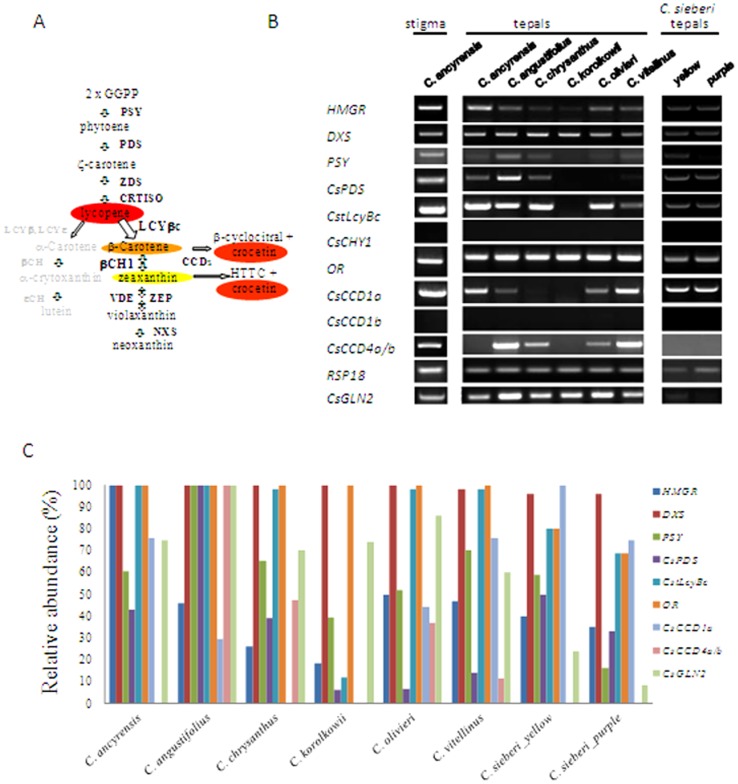
Figure 7. Expression analysis of selected genes involved in carotenoid metabolism.
A. Schematic carotenoid and apocarotenoid biosynthetic pathway in Crocus. Enzymatic reactions are represented by arrows. GGPP, geranyl geranyl diphosphate; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, z-carotene desaturase; CRTISO, carotene isomerase; LCYe, lycopene e-cyclase; LCYb, lycopene b-cyclase; bCH, b-carotene hydroxylase; eCH, e-carotene hydroxylase; ZEP, zeaxanthin epoxidase; VDE, violaxanthin de-epoxidase; NXS, neoxanthin synthase; CCD, carotenoid cleavage dioxygenase. B. RT-PCR analysis for expression of selected candidate genes related to the carotenoid metabolism. Equal amounts of total RNA were used in each reaction. The levels of constitutively expressed RPS18 coding gene were assayed as controls. The PCR products were separated by 1.5% (w/v) agarose gel electrophoresis and visualized by ethidium bromide staining. C. Relative transcript levels in tepals of the genes analysed by RT-PCR in B.