Abstract
Huanglongbing (HLB) is the most destructive disease that affects citrus worldwide. The disease has been associated with Candidatus Liberibacter. HLB diseased citrus plants develop a multitude of symptoms including zinc and copper deficiencies, blotchy mottle, corky veins, stunting, and twig dieback. Ca. L. asiaticus infection also seriously affects the roots. Previous study focused on gene expression of leaves and fruit to Ca. L. asiaticus infection. In this study, we compared the gene expression levels of stems and roots of healthy plants with those in Ca. L. asiaticus infected plants using microarrays. Affymetrix microarray analysis showed a total of 988 genes were significantly altered in expression, of which 885 were in the stems, and 111 in the roots. Of these, 551 and 56 were up-regulated, while 334 and 55 were down-regulated in the stem and root samples of HLB diseased trees compared to healthy plants, respectively. Dramatic differences in the transcriptional responses were observed between citrus stems and roots to Ca. L. asiaticus infection, with only 8 genes affected in both the roots and stems. The affected genes are involved in diverse cellular functions, including carbohydrate metabolism, cell wall biogenesis, biotic and abiotic stress responses, signaling and transcriptional factors, transportation, cell organization, protein modification and degradation, development, hormone signaling, metal handling, and redox. Microscopy analysis showed the depletion of starch in the roots of the infected plants but not in healthy plants. Collapse and thickening of cell walls were observed in HLB affected roots, but not as severe as in the stems. This study provides insight into the host response of the stems and roots to Ca. L. asiaticus infection.
Introduction
Huanglongbing (HLB), which is also known as citrus greening, is currently the most destructive disease that affects citrus plants. The disease has been associated with three species of a phloem-limited α-proteobacterium that is designated as Candidatus Liberibacter; i.e., Ca. L. asiaticus, Ca. L. africanus, and Ca. L. americanus [1]. In addition to graft transmission, Ca. L. asiaticus and Ca. L. americanus are transmitted by the Asian citrus psyllid Diaphorina citri, while Ca. L. africanus is transmitted by the African citrus psyllid Trioza erytreae [1]. While Ca. L. americanus and Ca. L. africanus are still geographically limited, Ca. L. asiaticus is widely distributed, being found in southeast Asia, the Indian subcontinent, the Arabian peninsula, the U.S.A., Cuba, Mexico, Jamaica, Honduras, and Brazil [1].
HLB diseased citrus plants develop a multitude of symptoms, some of which resemble zinc and copper deficiencies [1]. Visible symptoms include blotchy mottled, pale yellow and thin leaves, yellow shoots, corky veins, stunting and twig dieback. Affected fruits are small, lopsided, have aborted seeds, are acidic and bitter in taste, and ripen prematurely [1]. It has also been reported that Ca. L. asiaticus infection seriously affects the roots [2]. It has been observed that Ca. L. asiaticus infected trees are more adversely affected by extreme weathers than are healthy trees. Consequently, symptoms of stress, e.g., excessive leaf loss and premature fruit drop, occur in Ca. L. asiaticus infected trees. This stress intolerance is thought to result partially from a loss of fibrous root function. Recently, it was reported that HLB-diseased, four-year-old trees of Valencia sweet orange (Citrus sinensis) on citrumelo rootstock (Citrus paradisi x Poncirus trifoliata) showed a 31% and 38% reduction in fibrous root mass density for presymptomatic and symptomatic trees, respectively, compared to healthy trees. Similarly, HLB-diseased, three-year-old trees of Hamlin sweet orange (C. sinensis) on citrumelo rootstock showed a 30% and 37% reduction in fibrous root mass density for presymptomatic and symptomatic trees, respectively [3].
Anatomical aberrations in the Ca. L. asiaticus infected leaves compared to healthy leaves include the excessive accumulation of starch, callose depositions, phloem plugging, necrosis and collapse, the swelling of sieve elements and companion cell walls, and the disruption of chloroplast inner grana structures [1], [4]–[8]. Analyses of secondary metabolites in the Ca. L. asiaticus infected fruit compared to healthy fruit revealed increases in terpenes, hesperidin, naringenin, quercetin, limonin and nomilin agycones [9], [10]. Starch accumulates excessively only in aerial tissues, such as the phloem elements and vascular parenchyma in leaves and petioles, xylem parenchyma and spongy mesophyll cells and the phelloderm of stems, but it is depleted in roots [4], [6], [8]. While sucrose and fructose accumulates in both the leaf midribs and lobes, glucose accumulates only in the midribs [7]. In addition, the disruption of inner grana structures occurs only in leaf parts that are experiencing phloem plugging [5]. Rosales and Burns [11] also showed that within the same fruit, the indole-3-acetic acid concentration is higher in misshapen areas compared with those that are normal in shape. Previous studies indicate that Ca. L. asiaticus is distributed in bark tissue, leaf midrib, roots, and different floral and fruit parts, but not in endosperm and embryo, of infected citrus trees [12]. The leaves, stems, and roots play distinct roles in the photosynthesis and transportation of water, nutrients, etc. However, the effects of Ca. L. asiaticus infection on gene expression in the stems and roots remain to be elucidated despite the recent progress that has been made toward understanding the transcriptomes of leaves and fruit that are infected with Ca. L. asiaticus using either microarray or RNA-seq [4], [13]–[18].
Microarray analyses of infected sweet orange (Citrus sinensis) leaves revealed that Ca. L. asiaticus modulates a large cascade of molecular pathways [4], [13], [14], [17], to which some of the abovementioned phenotypes have been attributed. For example, in addition to callose deposition, the plugging of phloem elements has been linked to phloem protein 2 (PP2), which contributes to starch accumulation [4], [5], [13]. It was suggested that the excessive starch accumulation is partly due to the up-regulation of starch synthesis genes, such as ADP-glucose pyrophosphorylase (AGPase), starch synthase, granule-bound starch synthase (GBSS) and the starch debranching enzyme (SDE) [4], [13]. Recently, Fan et al. [7] reported the down-regulation of starch metabolism enzymes, such as transglucosidase and a maltose exporter. Furthermore, their work demonstrated an increase in cell wall invertase activity in HLB-affected citrus plants, which may lead to increases in sucrose and glucose. Not surprisingly, several biological processes differentially modulated in leaves during the symptomatic phase of Ca. L. americanus infection were similarly affected by Ca. L. asiaticus infection including induction of transcripts encoding zinc transporters [14], induction of transcripts encoding key enzymes involved in starch biosynthesis and repression of those related to starch breakdown, and induction of transcripts encoding P-proteins and repression of transcript encoding a salicylic acid-binding protein 3 (SABP3) [19].
Both microarray and RNA-seq techniques have been used to investigate the citrus fruit response to Ca. L. asiaticus infection [15], [18]. Liao and Burns [18] compared the transcriptomic changes associated with Ca. L. asiaticus infection in flavedo, vascular tissue, and juice vesicles from symptomatic, asymptomatic and healthy fruit based on microarray analysis. Their results indicated that many categories of metabolism including numerous genes involved in carbohydrate transport and metabolism were affected by HLB, but no category appeared to be specific to the disease. It was also suggested that mechanisms regulating development of HLB symptoms may result from the host disease response rather than being a direct consequence of carbohydrate starvation [18]. RNA-Seq was used to profile the transcriptome of citrus fruit response to Ca. L. asiaticus infection focusing on the peel [15]. Importantly, RNA-Seq can reveal rare and unknown transcripts and expression of genes unrepresented on the arrays, and avoid non-specific hybridization despite some drawbacks of RNA-Seq [15], [20]–[22]. It was shown that numerous pathways including those involved in photosynthesis, source-sink communication, sucrose and starch metabolism, and hormone synthesis and signaling were differentially regulated by Ca. L. asiaticus infection [15]. Proteomics analyses also helped us understand the host response of citrus to Ca. L. asiaticus infection [14], [23]. In this study, we compared the gene expression levels of stems, and roots in healthy Valencia sweet orange trees with those that were infected with Ca. L. asiaticus using microarrays. Microscopy analyses were also conducted to compare the stems and roots of healthy and HLB diseased citrus.
Results
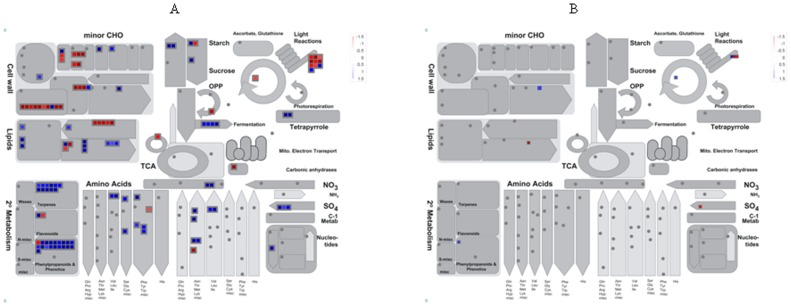
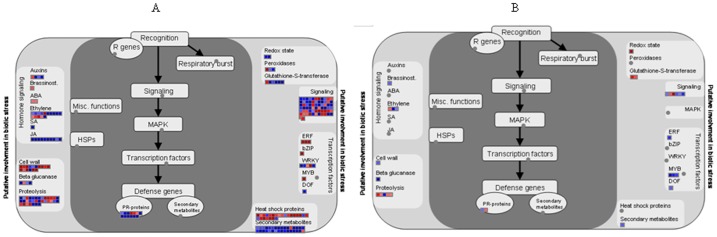
To understand how Ca. L. asiaticus infection affects stems and roots, Affymetrix microarray analysis was conducted. After removal of probe set-related redundancies, and at a P value of <0.05 and a log2 fold change (LFC) of ≥1.00 or ≤−1.00 as cutoff thresholds, a total of 988 genes showed significantly altered expression, of which 885 were in the stems, and 111 in the roots. Of these, 551 and 56 were up-regulated, while 334 and 55 were down-regulated in the stem and root samples, respectively (Fig. 1). Dramatic differences in the transcriptional responses were observed between the citrus stems and roots to Ca. L. asiaticus infection, with only 8 genes affected in both the roots and stems. For the identification of the processes and genes that were affected, the data was analyzed using the MapMan gene ontology system [24]. We found that the affected genes are involved in diverse cellular functions (Fig. 2, Fig. S1, Fig. S2), including carbohydrate metabolism (Fig. S3), cell wall biogenesis (Fig. 2), biotic and abiotic stress responses (Fig. 3), signaling and transcriptional factors (Fig. 3, Fig. 4, Fig. S2, Fig. S4), secondary metabolism (Fig. S5), phenylpropanoid pathway (Fig. S6), transportation, cell organization, protein modification and degradation, development, hormone signaling, metal handling, redox, and enzymatic activities (Fig. S1, Fig. S2). The proportions and types of genes that belonged to the various functional groups were different between roots and stems, with the most categories altered in the stems compared with the roots. The low similarities between the stem and root expression profiles indicate that Ca. L. asiaticus infection affects the stems, and roots differently. Based on previous knowledge regarding the responses of citrus plants to Ca. L. asiaticus infection, we focused further analyses on seven gene categories that are presumed to significantly contribute to the HLB disease symptoms caused by Ca. L. asiaticus infection.
Figure 1. Differential regulation of genes in the stems and roots of Valencia sweet orange (Citrus sinensis) by Ca. L. asiaticus infection.
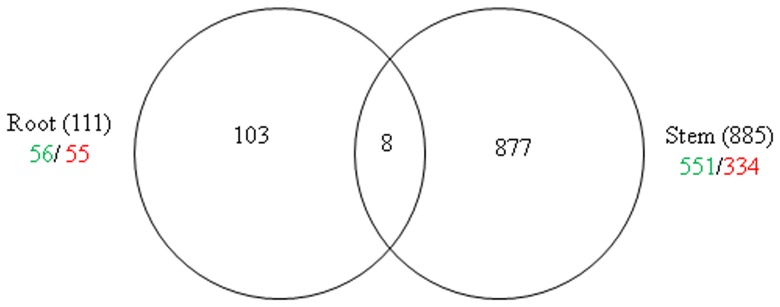
Figures in parentheses indicate total numbers of genes in the stems and in the roots. The numbers of significantly up-regulated genes are shown in green, and down-regulated are shown in red. A total of 998 genes showed significantly altered expression in the stems and roots.
Figure 2. Overview of metabolic pathways that are regulated by Ca. L. asiaticus infection in the stems and roots of Valencia sweet orange (Citrus sinensis).
A = stem and B = root. Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red.
Figure 3. Regulation of biotic stress-related gene pathways by Ca. L. asiaticus infection in the stems and roots of Valencia sweet orange (Citrus sinensis).
A = stem and B = root. Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red.
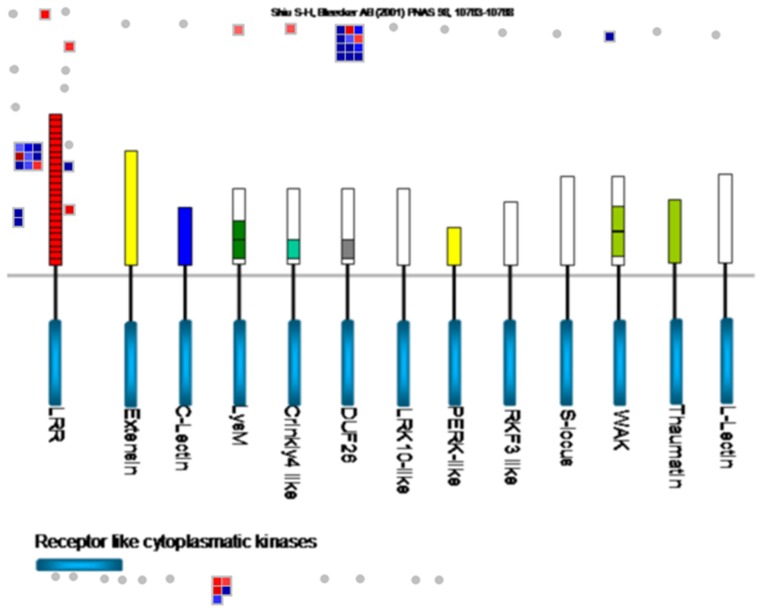
Figure 4. Regulation of receptor-like kinase (RLK) genes by Ca. L. asiaticus infection in the stems of Valencia sweet orange (Citrus sinensis).
Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red. Abbreviations/definitions: LRR, leucine-rich repeats; Extensin, RLK with extensin motif; LysM, RLKs with lysine motif; C-lectin, RLKs with lectin-like motifs; Crinkly4-like, RLKs with crinkly4-like domains; DUF26, domain of unknown function 26; LRK 10-like, RLK gene linked to Lr10 locus; L-lectin, RLKs with lectin-binding domains; PERK-like, proline-rich extensin-like kinase; S-locus, RLK with S-domain similar to S-locus glycoproteins; RKF3-like, receptor-like kinase in flowers 3; Thaumatin, RLK-like thaumatin protein; WAK, wall-associated kinase.
Starch and Sugar Metabolism
The induction of starch accumulation by Ca. L. asiaticus infection has been found to be in the leaves followed by stems but not in the roots [4], [6], [7]. Sucrose and glucose accumulation by Ca. L. asiaticus infection has also been found in the leaves [4], [7]. Therefore, we compared the expression of carbohydrate metabolism genes in the stems and the roots. In conformity to the above, the expression of the genes encoding enzymes and proteins that are involved in both major and minor carbohydrate metabolism was only affected in the stems, but not in the roots (Fig. 2, Fig. S3, Table 1). The transcription of genes encoding an ADP-glucose pyrophosphorylase large subunit 3 (APL3), which catalyzes the rate limiting step in starch synthesis [25], and a granule-bound starch synthase (GBSS), which initiates synthesis of starch granules, was up-regulated. Among the genes encoding starch-degrading and sucrose cleaving enzymes, the genes for alpha-amylase 1 (AMY1) and exocellular acid invertase 1 (Exinv1) were up-regulated, while those encoding beta-amylase 1 (BMY1) and a neutral invertase were down-regulated. Ca. L. asiaticus infection also repressed the transcription of minor carbohydrate metabolism associated genes encoding trehalose biosynthesis enzymes, trehalose-6-phosphate synthases (TPS2, TPS8, and TPS10) and two trehalose-6-phosphate phosphatases, but activated the gene for myo-inositol oxygenase (EC 1.13.99.1), which cleaves inositol to D-glucuronic acid (Fig. 2, Table 1).
Table 1. Differentially expressed genes related to carbohydrate metabolism and cell wall biogenesis in the stems and roots of Valencia sweet orange (Citrus sinensis) caused by Ca. L. asiaticus infection.
| Accession No. | Gene description | Log2 fold change | |
| Stem | Root | ||
| Starch synthesis | |||
| DN622894 | ADP-glucose pyrophosphorylase large subunit 3 (APL3) | 1.96 | |
| CB292132 | Granule-bound starch synthase (GBSS) | 1.82 | |
| Starch and sucrose degradation | |||
| CX046632 | Exocellular acid invertase 1 (Exinv1) | 2.43 | |
| CD575166 | Neutral invertase | −1.00 | |
| DN958063 | Alpha-amylase (AMY1) | 2.45 | |
| CF836730 | Beta-amylase (BMY1/BMY7) | −1.17 | |
| Minor carbohydrate metabolism | |||
| CN184547 | putative Myo-inisitol oxygenase | 1.91 | |
| CX046914 | Raffinose synthase family protein (din10) | 1.51 | |
| CX306291 | Stachyose synthase | −1.11 | |
| CD575394 | Trehalose-6-phosphate phosphatase (TPPA) | −1.31 | |
| CX636014 | Trehalose-6-phosphate phosphatase, putative | −1.64 | |
| CX309237 | Trehalose-6-phosphate synthase 2 (TPS2) | −1.21 | |
| CF831824 | Trehalose-6-phosphate synthase 8 (TPS8) | −1.63 | |
| CX306907 | Trehalose-6-phosphate synthase 10 (TPS10) | −1.83 | |
| Cell wall precursors | |||
| CK939533 | UDP-glucose 4-epimerase | −1.37 | |
| Cell wall proteins | |||
| CX638719 | Extensin | −1.41 | |
| CX076453 | Extensin-like protein | 1.22 | |
| CF417841 | Proline-rich protein (PRP4) | 1.09 | |
| CK938120 | Glycine-rich RNA-binding protein | 1.23 | |
| Cellulose synthesis | |||
| CF835773 | Cellulose synthase-like C7 (CSLC7) | −1.90 | |
| CV705580 | Cellulose synthase catalytic subunit-like protein | −1.72 | |
| DN618266 | Cellulose synthase-like protein D4 (CSLD4) | −1.44 | |
| CX045083 | Cellulose synthase isolog | 1.25 | |
| Cell wall modification | |||
| DN618428 | BURP domain containing protein | 2.01 | |
| CX665316 | Polygalacturonase-like protein | 1.10 | |
| CF835126 | Expansin-related protein 1 (EXP1) | 2.03 | |
| CK939135 | Syringolide-induced protein 19-1-5 | −2.67 | |
| CX296097 | putative Xyloglucan endotransglycosylase (XET) | −2.80 | |
| CV716643 | Xyloglucan endotransglycosylase 1 (XET) | −2.53 | |
Accession No. is a unique identifier of EST sequences from several citrus species and hybrids linked to the NCBI. LFC is the ratio of the expression level in the infected samples compared to the healthy trees. The ratio is the mean of 3 replicates. The annotation is according to the latest available BLASTx search at non-redundant protein database at the NCBI. Metabolic pathway grouping is based on the gene ontology in the MapMan program (Thimm et al., 2004).
Cell Wall Metabolism
Plant cell walls are composed of layers of cellulose microfibers embedded in a matrix of pectin and hemicellulose, plus some structural proteins [26]–[28]. Ca. L. asiaticus infection induced differential expression of multiple genes encoding enzymes and proteins involved in the synthesis, assembly, and modification of cell wall. The infection repressed the expression of genes encoding cellulose synthase-like D4 (CSLD4) and CSLC7 that mediate synthesis of β-1,4 linkages in the hemicellulose backbones, and a cellulose synthase catalytic subunit protein (Fig. 2, Table 1) [28], [29]. Mutations in cellulose synthesis associated genes lead to reduction in growth anisotropy, and display cell and organ swelling [27]. In rice, CSLD4 has been shown to be essential for normal cell-wall biosynthesis and plant growth [30]. Ca. L. asiaticus infection also repressed expression of a UDP-glucose 4-epimerase encoding gene. Isoforms of UDP-glucose 4-epimerase were reported to be involved in pollen development, influence cell wall galactose content, which was correlated with shoot growth, and affect root growth by changing galactant content [31]. This is consistent with the reduced shoot and root growth of the HLB diseased trees. Ca. L. asiaticus infection up-regulated the transcription of genes encoding structural protein components of the cell wall such as a proline-rich protein (PRP4), a glycine-rich RNA-binding protein and an extensin-like protein. Extensin is a structural protein that is involved in cell wall assembly [32]. The infection repressed expression of two genes encoding xyloglucan endotransglycosylase (XET) [33] which has been implicated in many aspects of cell wall biosynthesis including regulating wall expansion by cutting and rejoining xyloglucan to incorporate newly synthesized XG into the wall matrix [33] and in the wall degradation needed for fruit ripening [34]. Ca. L. asiaticus infection up-regulated genes encoding an expansin-related protein 1 (EXP1), and a BURP domain-containing protein. Expansin is a cell wall loosening protein that induces stress relaxation and extension of cell walls. Organ specific expression of EXP1 has been reported in hybrid aspen (Populus tremula x Populus tremuloides Michx) where it was highly expressed in stem tissues such as cambium/phloem but not in roots [35], [36]. In the root, only two genes were affected in expression by Ca. L. asiaticus infection including down-regulation of an extension gene and up-regulation of a gene encoding a polygalacturonase-like protein which hydrolyses galacturonic acid residues in cell walls (Fig. 2, Table 1).
Disease Resistance and Pathogenesis-related (PR) Genes
The expression of disease resistance associated genes belonging to the nucleotide binding site (NBS)- leucine-rich repeat (LRR) gene family that contain centrally located NBS, C-terminal LRR and amino-terminal TIR (Toll/interleukin-1 receptor-like) or CC (coiled-coil) domains were affected predominately in the stems compared to the roots by Ca. L. asiaticus infection (Table 2). In the stems, reduced transcription was observed for genes encoding a NBS-LRR-like protein cD7, a TMV N-like disease resistance protein that confers resistance to tobacco mosaic virus in tobacco [37], a putative TIR-NBS-LRR-class protein, and an inter-alpha-trypsin inhibitor heavy chain-related protein (Table 2). The genes encoding a CC-NB-LRR domain containing protein, a resistance protein candidate 2 (RGC2), and a disease resistance family protein SC0A belonging to the Cladosporium fulvum resistance protein Cf-9 (Hcr9) family, were however, up-regulated (Table 2). In the roots, while the expression of a gene encoding the CC-NB-LRR was repressed, expression of a gene encoding a NBS-LRR protein Hom-F was up-regulated by Ca. L. asiaticus infection. Also specifically in the stem, genes encoding miraculin-like proteins, which are soybean Kunitz-type trypsin inhibitors that specifically inhibit the activities of membrane-bound serine proteases [38], were up-regulated. Among PR proteins, only the transcription of a PR10-encoding gene was up-regulated in the stems but was un-affected in the roots (Table 2).
Table 2. Differentially expressed genes related to disease resistance and pathogenesis-related proteins in the stems and roots of Valencia sweet orange (Citrus sinensis) caused by Ca. L. asiaticus infection.
| Accession No. | Gene description | Log2 fold change | |
| Stem | Root | ||
| CX670605 | PR protein from class 10 | 1.03 | |
| CX674271 | Pathogenesis-related protein PR10A | 1.43 | |
| DN618117 | NBS-LRR-like protein cD7 | −1.32 | |
| CX674648 | TMV N-like disease resistance protein | −1.64 | |
| CX671207 | Inter-alpha-trypsin inhibitor heavy chain-related | −1.04 | |
| CB322091 | Disease resistance protein (TIR-NBS-LRR class) | −1.67 | |
| CD573651 | NBS-LRR type disease resistance protein Hom-F | 1.05 | |
| CX075252 | CC-NB-LRR protein | −1.06 | |
| DN796858 | CC-NB-LRR protein | 1.21 | |
| DN798196 | Resistance protein RGC2 | 1.28 | |
| CX641603 | Disease resistance protein, putative | 1.05 | |
| CX287601 | Disease resistance family protein, SC0A | 1.99 | |
| CK934670 | Miraculin-like protein 2 | 2.72 | |
| CX305265 | Miraculin-like protein 2 | 3.38 | |
| CX043805 | Miraculin-like protein 2 | 3.72 | |
| CX674833 | Miraculin-like protein 2 | 5.37 | |
| CX045518 | Miraculin-like protein 2 | 3.82 | |
| CF833881 | Miraculin-like protein 3 | 3.39 | |
Accession No. is a unique identifier of EST sequences from several citrus species and hybrids linked to the NCBI. LFC is the ratio of the expression level in the infected samples compared to the healthy trees. The ratio is the mean of 3 replicates. The annotation is according to the latest available BLASTx search at non-redundant protein database at the NCBI. Metabolic pathway grouping is based on the gene ontology in the MapMan program (Thimm et al., 2004).
Signaling Molecules and Receptor-like Kinases
The expression of several genes encoding receptor-like kinases (RLK) which regulate plant development and defense response was affected in the stems but not in the roots (Fig. 3, Table 3). Ca. L. asiaticus infection up-regulated expression of 12 plant defense genes (Fig. 3) such as homologs of Xa21, Hcr2-5D, Cf-5 and Cf-2.2, and a gene encoding a systemin receptor SR160 which was reported to induce systemic defense genes in wounded tomato plants [39]. Five development related genes including genes encoding a receptor protein kinase ERECTA and a receptor-like protein kinase 1 were down-regulated in the stems by Ca. L. asiaticus infection (Table 3). Ca. L. asiaticus infection up-regulated genes encoding FERONIA (FER) in the catharanthus roseus-like gene family [26] and WAK-like kinase (WLK2) in the wall-associated kinase gene family (Table 3). Cell wall-associated kinase (WAK) and WLKs family gene products which differ from RLKs by containing a cytoplasmic serine/threonine kinase domain and an extracellular region containing epidermal growth factor-like repeats regulate cell elongation and plant development as well as pathogen and heavy-metal tolerance [40].
Table 3. Differentially expressed genes related to signaling in the stems and roots of Valencia sweet orange (Citrus sinensis) caused by Ca. L. asiaticus infection.
| Accession No. | Gene description | Log2 fold change | ||||||
| Stem | Root | |||||||
| Crinkly like | ||||||||
| CV711750 | putative Protein kinase | −1.06 | ||||||
| Lysine motif | ||||||||
| CX293076 | putative Protein kinase | −1.01 | ||||||
| Wall associated kinase | ||||||||
| CD574549 | WAK-like kinase (WLK2) | 4.88 | ||||||
| Catharanthusroseus-like RLK | ||||||||
| DN622543 | FERONIA receptor-like kinase | 1.15 | ||||||
| MAP kinase | ||||||||
| CX073386 | MPK3 | −1.44 | ||||||
| Receptor like cytoplasmatic kinase | ||||||||
| CN184216 | protein Serine threonine kinase-like | −1.25 | ||||||
| CV707629 | Receptor protein kinase-like protein | −1.14 | ||||||
| CX670980 | putative Protein kinase | 1.58 | ||||||
| CV719828 | AT3G57730 | 1.16 | ||||||
| DUF 26 | ||||||||
| CN185603 | hypothetical protein F19K19.4 | −1.39 | ||||||
| CN192263 | AT1G01540 | −1.13 | ||||||
| DN625179 | Receptor protein kinase-like protein | 1.70 | ||||||
| CV710863 | putative Receptor-like protein kinase 4 RLK4 | 1.65 | ||||||
| CV706414 | Receptor protein kinase | 1.24 | ||||||
| CX674957 | hypothetical protein F20P5.3 | 1.93 | ||||||
| CX640171 | putative Serine threonine kinase | 1.60 | ||||||
| CX295060 | putative Serine threonine kinase | 1.07 | ||||||
| DN617645 | Receptor protein kinase | 2.43 | ||||||
| Leucine rich repeat | ||||||||
| CV707041 | probable Receptor kinase | −1.28 | ||||||
| CX675277 | putative Receptor protein kinase, ERECTA | −1.57 | ||||||
| CX043848 | putative Receptor protein kinase, ERECTA | −1.37 | ||||||
| CX671538 | Hcr2-5D | 2.37 | ||||||
| CX671347 | Cf-2.2 | 1.38 | ||||||
| CX641603 | Disease resistance protein, putative | 1.05 | ||||||
| DN624314 | Disease resistance protein (cf-5) | 1.84 | ||||||
| CX674240 | hypothetical protein | 1.98 | ||||||
| CX669184 | Leucine-rich repeat receptor-like kinase | 1.62 | ||||||
| CX072406 | OSJNBa0020J04.8 | −1.19 | ||||||
| CK937178 | OSJNBa0021F22.13 | 2.65 | ||||||
| CX674269 | putative Protein kinase Xa21 | 1.10 | ||||||
| CX669563 | putative Receptor kinase | 2.27 | ||||||
| CX045546 | putative Receptor-like protein kinase | 1.03 | ||||||
| DT214563 | putative Systemin receptor SR160 | 2.51 | ||||||
| DN623196 | Receptor protein kinase-like protein | 2.02 | ||||||
| CX669844 | Receptor-like protein kinase 1 | −1.18 | ||||||
| Calcium signaling | ||||||||
| DN622169 | Avr9/Cf-9 rapidly elicited protein | −1.16 | ||||||
| CK935615 | Calcium-binding allergen Ole e 8 | −1.59 | ||||||
| CN188862 | Calcium-binding EF-hand family protein-like | −1.91 | ||||||
| CK701540 | Calcium-binding pollen allergen, Polcalcin | 1.13 | ||||||
| CX047037 | EF-hand Ca2+-binding protein CCD1 | −1.49 | ||||||
| CX306727 | 39 kDa EF-Hand containing protein | −1.37 | ||||||
| DN795254 | putative Calmodulin-binding protein | −1.63 | ||||||
| CK935677 | putative Calmodulin-related protein | −1.62 | ||||||
| DN799085 | putative Calreticulin | 1.43 | ||||||
| CK933894 | unnamed protein product | −1.07 | ||||||
| Sugar and nutrient physiology | ||||||||
| CX294586 | Glutamate receptor family protein (GLR3.3) | 2.63 | ||||||
| CF837692 | Ligand gated channel-like protein | 1.10 | ||||||
| CV711517 | phosphate-induced protein 1(PHI-1)-like protein | 1.60 | ||||||
| CF838115 | Phosphate-responsive protein, putative | 1.15 | ||||||
| CX292843 | Photoassimilate-responsive protein precursor PAR-1a | −1.70 | 2.12 | |||||
| CV710376 | Ionotropic glutamate receptor homolog GLR4 | 1.03 | ||||||
| CX288476 | putative Ligand-gated ion channel subunit | 1.50 | ||||||
Accession No. is a unique identifier of EST sequences from several citrus species and hybrids linked to the NCBI. LFC is the ratio of the expression level in the infected samples compared to the healthy trees. The ratio is the mean of 3 replicates. The annotation is according to the latest available BLASTx search at non-redundant protein database at the NCBI. Metabolic pathway grouping is based on the gene ontology in the MapMan program (Thimm et al., 2004).
The expression of genes that are involved in transducing Ca2+ signals was altered by Ca. L. asiaticus infection. In the stems, expression of eight genes was repressed including genes encoding a calcium-dependent protein kinase Avr9/Cf-9 rapidly elicited protein (ACRE), a calcium-binding allergen Ole e 8, a calmodulin-binding protein, an EF-hand Ca2+-binding protein CCD1, and a 39 kDa EF-hand-containing protein but only one gene encoding calreticulin was up-regulated (Table 3). In the roots, only the transcription of a gene for calcium binding pollen allergen, polcalcin, was activated (Table 3). Among genes whose products are involved in sugar sensing and nutrient physiology, genes encoding a glutamate receptor (GLR) family protein GLR3.3, a phosphate-responsive protein, and a phosphate-induced 1 (PHI-1)-like protein were up-regulated in the stems (Table 3). The gene encoding a photoassimilate-responsive protein PAR-1a was repressed in stems, but up-regulated in the roots (Table 3). Ca. L. asiaticus infection also up-regulated the expression of a gene encoding an ionotropic glutamate receptor homolog GLR4 (Table 3).
Transcription Factors
Ca. L. asiaticus infection up-regulated the expression of a gene encoding an Arabidopsis response regulator 1 (ARR1) homolog but repressed the expression of a gene encoding a lateral organ boundaries domain protein 38 (LOB38) in the roots (Fig. 4, Fig. S4, Table 4). LOBs perform numerous developmental functions and are known to be expressed in boundaries of the shoot and at the base of secondary roots [41], [42]. In the stems, expression of four genes encoding AUX/IAA family proteins was repressed by Ca. L. asiaticus infection (Table 4). Meantime, expression of an auxin response factor (ARF10) gene was also repressed (Table 4). Genes encoding basic helix-loop-helix (bHLH) family proteins showed both patterns of regulation (4 repressed and 2 up-regulated). Noticeably, the transcription of a gene encoding a homeobox-leucine zipper protein was repressed in the stems but was up-regulated in the root (Table 4). Ca. L. asiaticus infection repressed the expression of three genes but induced one gene belonging to the APETALA2/Ethylene-responsive element binding family in the stems and in the roots respectively (Table 4). Expression of two MYB domain TF family genes including a tuber-specific and sucrose-responsive element binding factor gene was repressed in stems, whereas in the roots three genes in the same family including MYB68 were up-regulated (Table 4). Importantly, its homolog AtMYB68 is a root growth specific regulator, impacting overall plant development under unfavorable conditions [43]. MYB68 is a negative regulator of lignin deposition in Arabidopsis [43], and its down-regulation may affect lignification in Ca. L. asiaticus infected citrus. Ca. L. asiaticus infection only affected the expression of genes encoding proteins containing WRKY domain in the stems, wherein, it increased the expression of WRKY4 and WRKY23 but repressed that of WRKY30 (Table 4). Overexpression of WRKY4 enhanced susceptibility of Arabidopsis plants to P. syringae and suppressed PR1 gene expression [44]. The effect of up-regulation of WRKY4 in Ca. L. asiaticus infected citrus remains to be addressed.
Table 4. Differentially expressed genes related to transcriptional factors in the stems and roots of Valencia sweet orange (Citrus sinensis) caused by Ca. L. asiaticus infection.
| Accession No. | Gene description | Log2 fold change | |||||
| Stem | Root | ||||||
| AP2/EREBP, APETALA2/Ethylene-responsive element binding protein family | |||||||
| CF836437 | putative AP2 domain transcription factor | 1.27 | |||||
| CN185220 | Transcription factor TINY, putative | −2.56 | |||||
| CF831712 | AP2 domain transcription factor-like protein | −1.80 | |||||
| CK939963 | CaCBF1B | −2.12 | |||||
| ARF, Auxin Response Factor family | |||||||
| CX294853 | Auxin response factor 10 | −1.17 | |||||
| Arabidopsis response regulator (ARR) | |||||||
| BQ623949 | Response regulator1 (ARR1) | 1.19 | |||||
| AS2, Lateral Organ Boundaries Gene Family | |||||||
| CF834266 | LOB domain protein 38 | −1.59 | |||||
| Aux/IAA family | |||||||
| CD574798 | AUX/IAA protein | −2.21 | |||||
| DR404247 | AUX IAA protein | −1.18 | |||||
| CV704184 | AUX IAA protein | −1.07 | |||||
| CV708756 | Auxin-regulated protein | −1.46 | |||||
| bHLH, Basic Helix-Loop-Helix family | |||||||
| CX045057 | Basic helix-loop-helix (bHLH) family protein | 2.63 | |||||
| CK932760 | bHLH transcription factor, putative | −1.04 | |||||
| CX676159 | Helix-loop-helix DNA-binding protein-like | −1.60 | |||||
| CF506528 | putative bHLH transcription factor | −1.60 | |||||
| CX671833 | putative Transcription factor RAU1 | 1.39 | |||||
| CX044856 | unknown protein | −1.11 | |||||
| bZIP transcription factor family | |||||||
| CF507428 | bZIP transcription factor | −3.04 | |||||
| C2C2(Zn) CO-like, Constans-like zinc finger family | |||||||
| CX048448 | Zinc finger (B-box type) family protein | −1.62 | |||||
| C2C2(Zn) DOF zinc finger family | |||||||
| DN798578 | unknown protein | 1.49 | |||||
| CX640084 | unknown protein | 1.10 | |||||
| C2C2(Zn) GATA transcription factor family | |||||||
| CX049639 | putative protein | −1.08 | |||||
| Zinc finger family | |||||||
| DN622949 | Zinc finger protein, ZPT2-12 | −1.46 | |||||
| DN797852 | Zinc finger protein, ZPT2-12 | −1.52 | |||||
| CK939958 | Zinc finger (CCCH-type) family protein | −1.56 | |||||
| GRAS transcription factor family | |||||||
| CN186577 | Scarecrow gene regulator | −1.62 | |||||
| CB291917 | Scarecrow transcription factor family protein | −1.25 | |||||
| CX670106 | Scarecrow-like transcription factor 14 (SCL14) | −1.14 | |||||
| CV710984 | AP2 SCARECROW-like protein | 1.01 | |||||
| HB, Homeobox transcription factor family | |||||||
| DN798602 | Homeobox leucine zipper protein | −1.42 | 1.21 | ||||
| CV704865 | Homeobox 2 protein | 1.06 | |||||
| MYB domain transcription factor family | |||||||
| CX307046 | MYB-like DNA-binding domain protein | 1.68 | |||||
| CK936024 | MYB family transcription factor (MYB68) | 1.30 | |||||
| CF834992 | MYB-related transcription factor | −1.42 | 1.20 | ||||
| CF506570 | Tuber-specific and sucrose-responsive element binding factor | −2.69 | |||||
| WRKY domain transcription factor family | |||||||
| CK939682 | putative WRKY transcription factor 30 (WRKY30) | −1.91 | |||||
| AJ489051 | putative WRKY4 transcription factor | 3.16 | |||||
| CX050828 | putative WRKY-type DNA binding protein 23 (WRKY23) | 1.51 | |||||
| CX643787 | WRKY transcription factor | 1.32 | |||||
Accession No. is a unique identifier of EST sequences from several citrus species and hybrids linked to the NCBI. LFC is the ratio of the expression level in the infected samples compared to the healthy trees. The ratio is the mean of 3 replicates. The annotation is according to the latest available BLASTx search at non-redundant protein database at the NCBI. Metabolic pathway grouping is based on the gene ontology in the MapMan program (Thimm et al., 2004).
Zinc finger-containing transcriptional factors that are involved in plant developmental and defense showed both patterns of regulation in the stems (Table 4). Expression of the following genes were repressed in the stem: ZPT2-12, a member of the EPF-type zinc fingers that are involved in flower development and plant defense against pathogens [45]; a gene encoding a CCCH-type zinc finger family protein; and a gene of the C2C2(Zn)-GATA family that has been implicated in light-responsive gene transcription in Arabidopsis [46], and a gene of the C2C2(Zn)-CO-like family that regulates circadian clock-responsive genes [47]. Ca. L. asiaticus infection induced the expression of a gene of C2C2(Zn)-DOF family that are involved in the regulation of carbohydrate metabolism and plant defense [48], [49] in the stems and in the roots, respectively. With the exception of a gene encoding an AP2 SCARECROW (SCR)-like (SCL) protein, expression of three genes belonging to the GRAS transcription factor family was repressed in the stems, including SCL14 and a scarecrow gene regulator highly related to chitin-inducible gibberellin-responsive proteins (Table 4). SCR and SCL proteins are involved in GA-mediated regulation of plant growth [50]. Arabidopsis mutants of SCR showed aberrant root growth and impaired development in aerial plant organs such as hypocotyl, inflorescence stems and shoot axial organs [51], [52]. Some of these symptoms are similar to those induced by Ca. L. asiaticus.
Transportation
The expression of numerous genes that are involved in transportation of carbohydrates, drugs, water, oligosaccharides, and amino acids was altered by Ca. L. asiaticus infection in the stems (Table 5). Among the carbohydrate transporters, Ca. L. asiaticus infection repressed the expression of a sugar transporter gene CiSUT1 but increased the transcription of genes encoding a hexose transporter 6 (HEX6) and a sorbitol transporter in the stems. In the roots, only the transcription of a sorbitol transporter gene was repressed (Table 5). Expression of three genes encoding amino acid transporters was up-regulated whereas one was down-regulated in the stems. Only one gene whose expression was up-regulated, encoding a putative amino acid transporter highly related (e-value, 4e-41) to the amino acid/polyamine transporter II of Medicago truncatula, was identified in the roots. Ca. L. asiaticus is auxotrophic for at least five amino acids [53]. It remains to be determined whether the up-regulation of such transporter genes results from the metabolic incapacity on certain amino acids of Ca. L. asiaticus.
Table 5. Differentially expressed genes related to transportation in the stems and roots of Valencia sweet orange (Citrus sinensis) caused by Ca. L. asiaticus infection.
| Accession No. | Gene description | Log2 fold change | |
| Stem | Root | ||
| Sugar transporter | |||
| CK936504 | putative Sorbitol transporter | −1.41 | |
| DN958552 | Sorbitol transporter | 1.25 | |
| CB292174 | Hexose transporter, putative | 1.28 | |
| CX665499 | Hexose transport protein HEX6 | 1.28 | |
| CK936487 | Citrus sucrose transporter 1 (CiSUT1) | −1.16 | |
| Amino acid transporters | |||
| CF418599 | Amino acid transporter family protein | 1.26 | |
| CV713475 | putative Cationic amino acid transporter | −1.02 | |
| CX298643 | Amino acid transporter family protein | 1.16 | |
| CF832082 | Cationic amino acid transporter 5 (CAT5) | 1.62 | |
| CK935365 | putative Amino acid transport protein | 1.45 | |
| Metabolite transporters | |||
| CK939426 | Mitochondrial substrate carrier family protein | −2.05 | |
| Major Intrinsic Proteins (NIP, PIP, SIP) | |||
| CX638658 | Major intrinsic protein | −2.47 | |
| DN958192 | NOD26-like intrinsic protein 6;1 | 2.71 | |
| CX644460 | putative Aquaporin (PIP1-1) | −1.50 | |
| CV719411 | Plasma membrane aquaporin (PIP1A) | −1.03 | |
| AU300362 | Putative aquaporin (PIP1-3) | −1.38 | |
| CK933592 | Small basic membrane integral protein 1A (SIP1A) | 1.68 | |
| Metal transport | |||
| CX297247 | Metal tolerance protein B1 (MTPB1) | −1.02 | |
| DN619440 | putative Zinc transporter | 1.48 | |
| CB290596 | Zinc transporter protein, ZIP1 | 3.76 | |
| Peptides and nucleotide transport | |||
| CN189143 | putative Proton-dependent oligopeptide transport (POT) family | 1.51 | |
| BQ622927 | Yellow stripe like 5 (YSL5) | 1.51 | |
| DN622714 | Nitrate transporter 1 | 1.57 | |
| CX305691 | Nitrate transporter NRT1-2 | 3.72 | |
| Phosphate and potasium transport | |||
| CX044970 | Phosphate transporter 3 | 1.29 | |
| CX047721 | Potassium channel tetramerisation domain-containing protein | −1.08 | |
| Vesicle transport and secretory pathways | |||
| CX293833 | putative Coatomer protein complex, subunit beta 2 | 1.74 | |
| CV708721 | unknown protein | −1.66 | |
| CX295788 | CAO | −1.18 | |
| CV885826 | VAMP family protein | −1.03 | |
| CF653150 | VAMP family protein | −1.09 | |
| CF837201 | putative RNA-binding protein | −1.49 | |
| ABC transporters and multidrug resistance systems | |||
| CB292087 | putative ABC transporter family protein | 3.11 | −1.80 |
| CX668058 | P-glycoprotein-like protein | 1.28 | |
| CX293439 | putative MRP-like ABC transporter | 1.11 | |
| Miscellaneous | |||
| CX671045 | putative protein | −1.04 | |
| CV709319 | Purine permease 1 (PUP1) | 1.37 | |
| CV707033 | Urea transporter DUR3 | 1.61 | |
| CF828901 | putative MATE efflux protein family protein | 1.18 | |
| CX674953 | MATE efflux family protein | 1.31 | |
| CV706445 | Chloride channel-like (CLC) protein, putative | 1.22 | |
| CX670490 | Nucleobase ascorbate transporter 12 (NAT12) | 1.40 | |
Accession No. is a unique identifier of EST sequences from several citrus species and hybrids linked to the NCBI. LFC is the ratio of the expression level in the infected samples compared to the healthy trees. The ratio is the mean of 3 replicates. The annotation is according to the latest available BLASTx search at non-redundant protein database at the NCBI. Metabolic pathway grouping is based on the gene ontology in the MapMan program (Thimm et al., 2004).
Among metal transporter genes, Ca. L. asiaticus infection activated the expression of genes encoding two zinc transporters (such as ZIP1), but repressed that of a metal tolerance protein B1 (MTPB1) (Table 5). In addition, expression of genes encoding transporters that move peptides, oligopeptides, and ions across membranes, including a POT family gene, NRT1 and YSL5, was up-regulated in the stems but not in the roots (Table 5). Expression of genes whose products perform various functions such as PUP1 (purine permease 1) which is involved in the uptake and transportation of cytokinins [54]; urea transporter DUR3 involved in acquisition, transportation, and utilization of urea [55]; and a chloride channel-like (CLC) protein which moves chloride ions across membranes, and two multidrug and toxic compound extrusion (MATE) efflux family proteins was also up-regulated (Table 5). The expression of a gene encoding a nucleobase ascorbate transporter 12 (NAT12) was up-regulated in the roots (Table 5).
Secondary Metabolic Pathway
The expression of genes encoding proteins and enzymes that synthesize flavonoids, isoprenoids, phenylpropanoids and lignin was mostly up-regulated by Ca. L. asiaticus infection (Table 6). Only the expression of one gene encoding a naringenin-chalcone synthase 4 (CHS4) was repressed in the stems. Ca. L. asiaticus infection induced the transcription of 10 genes in the isoprenoid metabolic pathway such as genes encoding a 1-deoxyxylulose 5-phosphate synthase which catalyses the rate-limiting step in the production of isopentenyl diphosphate, the main precursor of all isoprenoids, a beta-amyrin synthase, a geranyl diphosphate synthase small subunit, a homogentisate geranylgeranyl transferase, two linalool synthases, and a gamma-terpinene synthase (Table 6). Expression of genes involved in the phenylpropanoid pathway were similarly up-regulated including genes encoding phenylalanine ammonia lyase (PAL; EC 4.3.1.5), a key enzyme that converts P-phenylalanine to trans-cinnamic acid, a precursor for various phenylpropanoids [56], a hydroxycinnamoyl transferase (HCT; EC 2.3.1.133), which catalyzes the conversion of p-coumaroyl CoA or cafeoyl CoA with shikimic acid to p-coumaroyl shikimate or caffeoyl shikimate; as well as a 10-hydroxygeraniol oxidoreductase, a caffeic acid O-methyltransferase II, the 4-coumarate-CoA ligase-like protein and catechol O-methyltransferase (Fig. S6, Table 6). The expression of only one gene encoding a transferase family protein closely related to anthranilate N-benzoyltransferase involved in phenylpropanoids pathway was induced in the roots (Table 6).
Table 6. Differentially expressed genes related to secondary metabolism in the stems and roots of Valencia sweet orange (Citrus sinensis) caused by Ca. L. asiaticus infection.
| Accession No. | Gene description | Log2 fold change | |
| Stem | Root | ||
| Flavonoids | |||
| CX672036 | putative Flavanone 3-hydroxylase | 2.21 | |
| CX045954 | Naringenin-chalcone synthase 4 | −1.14 | |
| Isoprenoids | |||
| CX302245 | 1-deoxyxylulose 5-phosphate synthase | 1.15 | |
| CF838068 | Beta-amyrin synthase | 1.58 | |
| CX290062 | Geranyl diphosphate synthase small subunit | 2.70 | |
| CX667086 | GGPP synthase | 1.28 | |
| CX043706 | HMG-CoA synthase 2 | 1.22 | |
| CX665915 | Homogentisate geranylgeranyl transferase | 2.14 | |
| CV886253 | Linalool synthase | 2.51 | |
| CV885575 | Linalool synthase | 3.41 | |
| CX045048 | Gamma-terpinene synthase | 3.75 | |
| CX671596 | Acetyltranferase-like protein | 1.29 | |
| CK937255 | Acetyltranferase-like protein | −1.23 | |
| Phenylpropanoids/lignin biosynthesis | |||
| CX671596 | Acetyltranferase-like protein | 1.29 | |
| CX044256 | Hydroxycinnamoyl transferase | 2.12 | |
| CF830793 | Transferase family protein | 1.47 | |
| CX663894 | Transferase family protein | 1.11 | |
| CV884611 | 4-coumarate-CoA ligase-like protein | 1.33 | |
| CV705977 | 4-coumarate-CoA ligase-like protein | 1.14 | |
| CB291954 | 10-hydroxygeraniol oxidoreductase | 2.08 | |
| DN622570 | putative Orcinol O-methyltransferase | 3.86 | |
| CX043719 | Caffeic acid O-methyltransferase II | 1.73 | |
| CX303448 | Catechol O-methyltransferase | 1.34 | |
| CX670983 | Catechol O-methyltransferase | 2.53 | |
| CX302017 | O-methyltransferase | 1.34 | |
| CX643181 | Phenylalanine ammonia-lyase 2 (PAL2) | 3.62 | |
| CF835217 | Phenylalanine-ammonia lyase | 2.55 | |
| Amino acid synthesis | |||
| DN620167 | Prephenate dehydrogenase family protein | 1.24 | |
| CF834506 | Prephenate dehydratase family protein | 1.10 | |
| CV706063 | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase | 1.75 | |
Accession No. is a unique identifier of EST sequences from several citrus species and hybrids linked to the NCBI. LFC is the ratio of the expression level in the infected samples compared to the healthy trees. The ratio is the mean of 3 replicates. The annotation is according to the latest available BLASTx search at non-redundant protein database at the NCBI. Metabolic pathway grouping is based on the gene ontology in the MapMan program (Thimm et al., 2004).
Validation of Differentially Expressed Genes by Quantitative Reverse Transcription PCR (qRT-PCR)
To confirm the validity of the microarray experiment, qRT-PCR assays were performed. Seven genes encoding the SBIP1A, PRP4, CAT5, PAR-1a, GLR4, the tuber-specific and sucrose-responsive element binding factor (TSF), and ZIP1 were chosen for this confirmation (Table 6). These genes were chosen because they showed distinct patterns of expression. Common patterns of expression changes were observed by the two methods, indicating the reliability of the microarray data (Table 7).
Table 7. Comparison of expression of selected genes in the stems and roots of Valencia sweet orange (Citrus sinensis) to Ca. L. asiaticus infection based on quantitative reverse transcription-PCR and microarray analyses.
| Gene product description | Microarray | qRT-PCRa | |||
| Stems | Roots | Stems | Roots | ||
| CK933592 | Small basic membrane integral protein | 1.68 | 3.65 | ||
| CF417841 | Proline-rich protein PRP4 | 1.09 | 1.40 | ||
| CF832082 | Cationic amino acid transporter 5 | 1.62 | 2.13 | ||
| CX292843 | Photoassimilate-responsive protein PAR-1a | 2.20 | 4.96 | ||
| CV710376 | Ionotropic glutamate receptor homolog GLR4 | 1.03 | 2.42 | ||
| CF506570 | Tuber-specific and sucrose-responsive element binding factor | −2.69 | −2.93 | ||
| CB290596 | Zinc transporter protein ZIP1 | 3.75 | 2.56 | ||
Fold change value calculated according to the method of Livak and Schmittgen (2001). The following primers designed using the PrimerQuestSM program (Integrated DNA Technologies, Inc) were used, CK933592: TGCAGTGTTGACATCTCTGTGGGT/ACTGGTAACAGGGCTTCAACTCCA;
CF417841: ATCAGGCACTCCATGTCCAGCTTA/GTATAAGCAGCGGTTGAAGCAGCA; CF832082: TGTCGCGCTTCTTGTGAGGAGATA/TCCCAAGAACCAAAGAGGAACGGT; CX292843: TATGCGAAGCACAAGGGAAAGGTG/AAAGTTCCGGTTATGTGGCACCGGC;
CF506570: ACCAGGCTTGTCGAACGATATGGA/TGCATGATTTCCCAGACCTTCCCT;
CV710376: TGCTTGGCTTCAGGTCAGGGAATA/AGCCTCCGAACGTGTAAGTGTTGA;
CB290596: AAGGGATATTCAACGCAGCAGCAG/ACAAGGACATGCAACCAGCTCCTA.
Microscopy Analyses of Stems and Roots Infected by Ca. L. asiaticus
Light microscopy analyses of the stems of healthy and Ca. L. asiaticus infected plants were shown in Fig. 5. Phloem fibers were shown on the outer boundary of the phloem layer, with cambium and xylem on the inner boundary (Fig. 5). The difference between the stems from the healthy and Ca. L. asiaticus infected plants observed with light microscope was the increased amount of cell layers in the phloem and the increased thickness of the cell walls of some of the cells in this area (arrows). When observed at higher magnification (TEM), the following were observed for the stems of Ca. L. asiaticus infected plants: a swelling in the middle lamella (Fig. 5 D, E) in some areas and, in others, the increase in thickness and collapse of cell walls (Fig. 5 E, F) that appeared to be those of the sieve elements (SE) and companion cells (CC). This collapse observed in the stem was similar to, but not as severe as, that reported in petioles and mid-ribs of leaves affected by HLB. There was also typically more starch (S) in the HLB affected tissues (Fig. 5 C, D, Fig. 6 E, F) compared to the healthy.
Figure 5. Microscopic analyses of stems of healthy and HLB affected Valencia sweet orange.
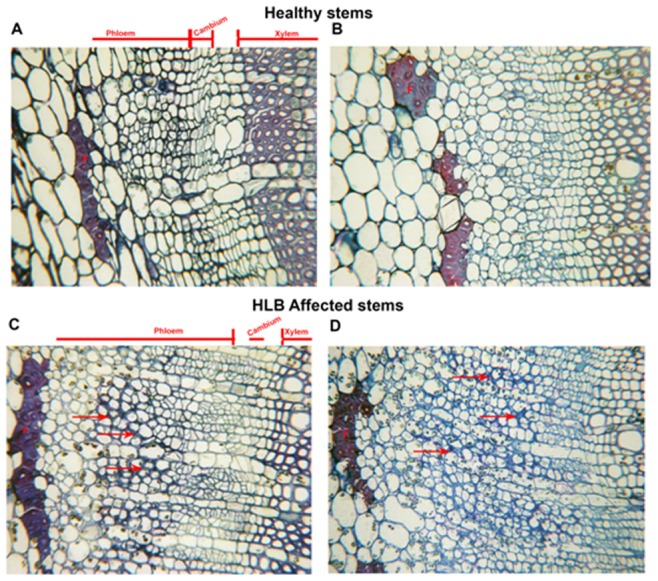
A, B. Light microscopy of cross-sections of healthy young stems showing the phloem, cambium and xylem cells. F-Phloem fibers. C,D Cross section of HLB affected young stems showing greater thickness of the phloem layer compared to the healthy. Arrows point to thickened cell walls.
Figure 6. Microscopic analyses of stems of healthy and HLB affected Valencia sweet orange.
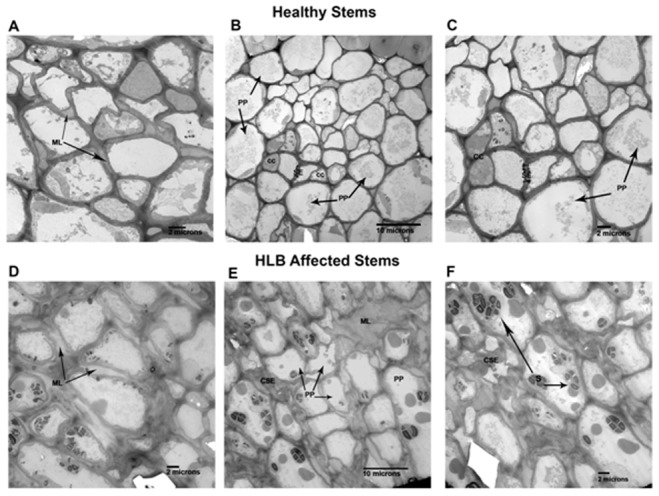
A, B, C are electron microscopy of cross- sections of healthy young stems. Arrows in A point to normal middle lamella found in healthy stem phloem. B is a lower magnification of healthy phloem showing normal companion cells (CC) on either side of sieve elements (SE) which are surrounded by phloem parenchyma cells (PP). C is an enlargement of the sieve element (SE) area pictured in B. D, E, F are cross-sections of HLB affected young stems. Arrows in D point to swollen middle lamella comparable to dark blue stained walls shown in Fig. 5. C, D. E shows phloem in affected stem comparable in magnification to B showing collapsed sieve element (CSE) surrounded by normal looking phloem parenchyma (PP) some of which contain starch (S). In upper right of micrograph is area showing swollen middle lamella (ML). F is an enlargement of area in lower left corner of E, showing collapsed sieve element (CSE) and parenchyma cells containing starch (S).
The comparison of healthy and HLB affected roots did not reveal as marked a contrast as in the stem (Fig. 7). The major differences included more starch (S) found in the roots of healthy plants (Fig. 7 A, B) compared to the HLB affected root (Fig. 8 C, D). There was not an increase in the development of phloem cells in the HLB roots as there was in the HLB stem tissue (Fig. 6). At the light microscope level, the collapse and thickening of cell walls of HLB affected roots (Fig. 7 C, D arrows) were not as severe as in the stems (Fig. 5). At the higher magnification (TEM), thickening of the middle lamella (ML) (Fig. 4, E) and thickening and collapse of the sieve elements (CSE) (Fig. 4 F) were also observed in the HLB affected roots. Another difference observed was the apparent greater density of the cytoplasm (Fig. 8 D) in the HLB affected phloem compared to that in the cells of the healthy plant (Fig. 8 A, C). Nuclei (N) and other organelles (Fig. 8 E, F) in the phloem parenchyma (PP) and companion cells (CC) in HLB affected roots were more prominent than in the healthy roots.
Figure 7. Microscopic analyses of roots of healthy and HLB affected Valencia sweet orange on citrumelo rootstock (Citrus paradisi x Poncirus trifoliata).
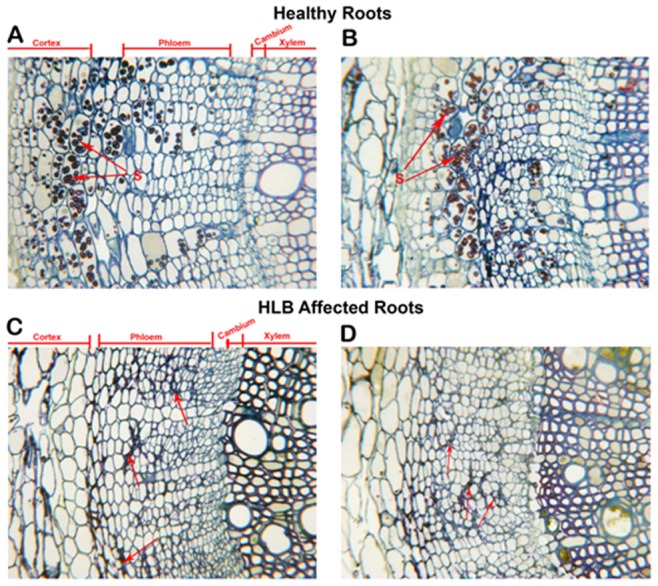
A, B. Light microscopy of cross-sections of healthy fibrous roots showing secondary thickening. The cortex and some phloem parenchyma cells are shown containing starch (S). The cortex, phloem, cambium and xylem areas are delineated by lines. C, D are cross-sections of HLB affected roots. Note that the phloem layers of these roots are comparable in thickness to those of healthy above. The arrows point to collapse, thickened cell walls.
Figure 8. Microscopic analyses of roots of healthy and HLB affected Valencia sweet orange on citrumelo rootstock (Citrus paradisi x Poncirus trifoliata).
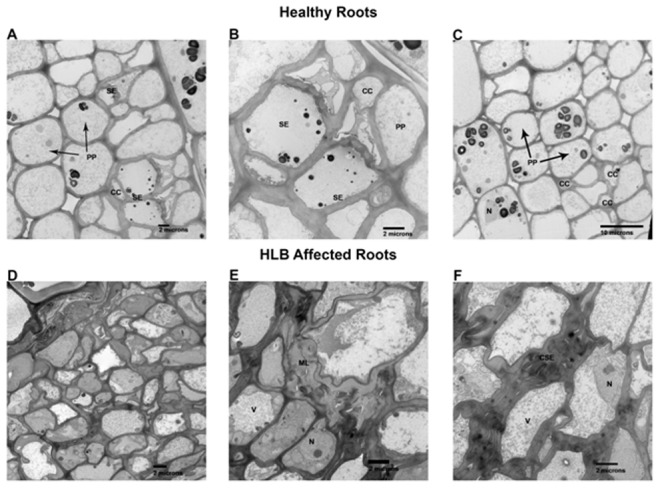
A–D. Electron microscopy of healthy fibrous roots. A & C are relative low magnifications showing size and shape of normal phloem parenchyma (PP), sieve elements (SE) and companion cells (CC). B shows a higher magnification of same areas enhancing the sieve element areas. D–F are HLB affected roots. D is a low magnification comparable to A & C above showing the enriched cytoplasmic contents of these cells compared to the healthy above. E shows enlarged middle lamellas (ML). F shows the collapsed sieve elements (CSE). N, prominent nucleus and V, vacuole.
Discussion
Dramatic differences in the transcriptional responses were observed between the citrus stems and roots to Ca. L. asiaticus infection (Fig. 1). Overall, 885, and 111 genes were regulated in the stems, and roots, respectively, by the infection, with only 8 genes overlapping (Fig. 1). In contrast to the low similarity between the expression profiles of the stems and roots, the expression profile of the stem to Ca. L. asiaticus infection is very similar to the leaf [4], [13]–[15], [17], [57]. For example, when we compared our data with the leaf expression profile [4], 291 genes were regulated in both the stems and the leaves compared to the 8 genes shared between the stems and roots (Fig. 1). The low similarity among the stem and root expression profiles might result from that Ca. L. asiaticus affects the stems and roots differently due to the distinct functions of the stems and roots, and the different physiological changes caused by the pathogen. Depletion of starch in the roots while excessive starch accumulation in the stems have been reported in Ca. L. asiaticus infected plants previously [4], [6], [8] as well as in this study (Fig. 5, Fig. 7). We could not rule out other possibilities that might contribute to the expression differences between the roots and the stems such as the stems and the roots are in different stages of infection since the plants were graft-transmitted. The collapse and thickening of cell walls in HLB affected roots (Fig. 7 C, D) were not as severe as in the stems (Fig. 5) suggesting that roots were in earlier stage of infection compared to the stems. The early stage of infection of the roots by Ca. L. asiaticus probably contributes to that only 111 genes were significantly affected in expression in roots.
The overall expression pattern of the stems to Ca. L. asiaticus infection shares high similarity with that of the leaves. High similarity between the expression profiles of the stems and the leaves might be due to that they share some common functions and have some similar phenotypes to Ca. L. asiaticus infection. The accumulation of starch, sucrose and glucose as induced by Ca. L. asiaticus has been found to be highly concentrated in both the leaves and stems (Fig. 5, Fig. 7) [4], [6], [7]. Collapse of sieve elements of the stems (Fig. 5) was similar to, but not as severe as the leaves (data not shown). The expression pattern of the stems to Ca. L. asiaticus infection suggests a very similar virulence mechanism of the pathogen as we learned from the leaf expression pattern including affecting carbohydrate metabolism, phloem blockage and aberrations, cell wall, plant defense, and hormones [58]. The excessive accumulation of starch in the stems (Fig. 7) is due to the similar mechanism as that of the leaves. This accumulation has been attributed to callose deposition, sieve pore plugging by the PP2 protein, and phloem necrosis and collapse [4], [13]–[15], [17], [57]. In addition to the above, other factors, including limited starch breakdown due to the down-regulation of starch-degrading genes, such as amylases and disproportionating enzyme 2 (DPE2), also contribute to the starch accumulation. Starch is degraded by AMY to maltose, and by BMY (BAM) together with DPEs to maltose and glucose. Ca. L. asiaticus infection increased the expression of AMY1 but repressed that of BMY1 (Table 1). While AMYs play minor roles, BMYs are indispensable for the breakdown because they catalyze the rate-limiting step [59]. Fan et al. [7] linked the accumulation of soluble sugars, such as glucose and fructose, to the up-regulation of cell wall-bound invertases. In our study, expression of the gene encoding Exinv1 was also up-regulated in the stems (Table 1).
Both the expression patterns of the stems and leaves suggest that Ca. L. asiaticus infection resemble a susceptible interaction between citrus and the pathogen [4], [13]–[15], [17], [57]. Ca. L. asiaticus infection repressed the expression of multiple genes that contain NBS-LRR domains (Table 2). NBS-LRR proteins belong to a major class of disease resistance molecules that recognize pathogenic effectors or plant proteins that are targeted by effectors, and many map to the R (resistance) gene loci [60]. They activate defense signaling [60]. The down-regulated genes encode homologs of the NBS-LRR-like protein cD7, the Tobacco mosaic virus (TMV) N-like disease resistance protein [37], and a putative CC-NBS-LRR gene that is related (e-value of 2e-43) to the potato (Solanum tuberosum) CC-NB-LRR class gene that was rapidly activated by an incompatible race of Phytophthora infestans [61]. The down-regulation of these genes indicates that Ca. L. asiaticus infection represses the defense response of citrus, leading to the susceptibility. Interestingly, our previous study indicated that Ca. L. asiaticus encodes a functional salicylate hydroxylase (SahA) that converts SA into catechol, which does not induce resistance [58]. SA has been proven to be an endogenous resistance signal, and its derivative MeSA is one of the signals for systemic acquired resistance (SAR). The accumulation of SA and its derivatives are necessary for SAR because it has been shown that plants that are unable to accumulate SA through the transgenic expression of a bacterial salicylate hydroxylase (NahG) that metabolizes SA into catechol are deficient in SAR. The expression of the bacterial salicylate hydroxylase enzyme in planta led to decreased SA levels, the failed development of SAR, or the decreased expression of the PR genes and heightened susceptibilities to both virulent and avirulent pathogens [62], [63]. On the other hand, Ca. L. asiaticus infection also activated the transcription of numerous defense related genes including genes encoding PR10, CC-NB-LRR protein, and disease resistance family protein SC0A (Table 2). However, the induction of plant defense related genes is incapable of preventing the establishment of Ca. L. asiaticus in the phloem. Collectively, the expression pattern of plant defense related genes to Ca. L. asiaticus infection resemble that of the susceptible plant pathogen interactions [64], [65].
We observed certain expression pattern associated with the stems that was not identified previously with the leaves. For example, Ca. L. asiaticus infection repressed expression of genes encoding the pore-forming aquaporins PIP1A, PIP1-1, PIP1-3 while induced expression of SIP1A gene, that belong to the MIP gene family, in the stems (Table 5), where the long distance movement of substances is common. Aquaporins are important molecules in plant physiology because they facilitate the transportation of water and other small, uncharged solutes, such as glycerol, CO2, ammonia and urea, from sources, such as roots and leaves, through the stem to other plant parts [66]–[68]. Their repression in citrus by Ca. L. asiaticus infection can deprive affected plants of water and some essential nutrients. Additionally, Ca. L. asiaticus infection caused dramatic effect on the phloem (Fig. 5, Fig. 6) in the stems as the major transportation pathway, which might contribute to the disease symptoms in the roots (Fig. 7, Fig. 8). On the other hand, the detrimental effect of HLB on roots will have negative feedback effect on the aerial parts of the plant.
Interestingly, swelling in the middle lamella was observed in both Ca. L. asiaticus infected stems and roots, which has not been reported previously (Fig. 6, Fig. 8 ). Swelling in the middle lamella was also observed in the presymptomatic leaf of Ca. L. asiaticus infected citrus [8]. The adjacent cells were separate from each other as observed in Fig. 6 and Fig. 8 which might affect nutrient transport among the neighboring cells via plasmodesmata, leading to eventual cell death. However, Ca. L. asiaticus does not encode plant cell-wall degradation enzymes such as cellulases, pectinases, xylanases, or endoglucanases [53]. Ca. L. asiaticus might indirectly affect the middle lamella by interfering with the plant cell wall related enzymes encoded by citrus such as rhamnogalacturonate lyase which are known to be regulated by plant hormones such as abscisic acid (ABA) and auxins [69]. Ca. L. asiaticus was reported to affect plant hormones including IAA and ABA in the fruit [11]. A detailed analysis of hormones in different tissues at different infection stages is required to test this hypothesis in the future. It remains to be determined how Ca. L. asiaticus causes the anatomical changes in middle lamella and how those changes affect HLB symptom development.
In conclusion, we analyzed the host response of citrus stems and roots to Ca. L. asiaticus infection. Dramatic differences in the transcriptional responses were observed between the citrus stems and roots to Ca. L. asiaticus infection. Microscopy analyses indicated that Ca. L. asiaticus infection significantly affects the starch accumulation, and sieve elements of the stems or roots. Understanding how to reduce the adverse effect of Ca. L. asiaticus infection on the leaves, stems and roots is critical to manage HLB.
Materials and Methods
Source of Plant Materials
Two-year-old Valencia sweet orange (C. sinensis) on rootstock Swingle citrumelo (Citrus paradisi. Macf. × Poncirus trifoliata [L.] Raf.) plants used in this study were graft-inoculated with budwood from Ca. L. asiaticus infected citrus trees and maintained in a USDA-APHIS/CDC-approved secured greenhouse. The inoculated plants that were used in this experiment were Ca. L. asiaticus-free before the graft inoculations, as shown by PCR and Q-PCR tests using specific primers [12]. Stem and root samples were obtained from three HLB symptomatic trees and three healthy control trees of similar size and from similar positions approximately 16 months after inoculation. The presence of Ca. L. asiaticus in the plants was confirmed using both conventional and quantitative PCR as described previously [12].
Microarray Analysis
Total RNA from the stems and roots were isolated from freshly obtained samples using the RNeasy Plant Mini Kit and treated with DNase (Qiagen, Valencia, CA). Root samples were prepared by excising small pieces from lateral roots and were frozen in liquid nitrogen before RNA purification. Stem pieces were harvested from young stems bearing symptomatic leaves by peeling off the bark and phloem together. For the softer stem parts, the whole stem was cut into smaller pieces and processed, as described above for the roots. The samples were grinded in liquid nitrogen with a mortar and pestle, the powder was rapidly suspended in RLT buffer (Qiagen, Valencia, CA) that was supplemented with 1% mercaptoethanol, and the solution was processed using the RNeasy Plant Mini Kit according to the manufacturer’s instructions. The quality of RNA was checked using the NanoDrop™ 1000 spectrophotometer (NanoDrop Technologies, Inc.), and only samples with A260/280 and A260/230 nm ratios of ∼2.0 were selected. The integrity of the total RNA was further determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Microarray experiments were carried out at the Gene Expression Core Facility of the Interdisciplinary Center for Biotechnology Research at the University of Florida. Data analyses were conducted as described previously [4]. Briefly, the Affymetrix GeneChip was used for microarray analysis. The GeneChip Citrus Genome Array contains 30,171 probe sets representing up to 33,879 citrus transcripts based on EST sequences obtained from several citrus species and citrus hybrids. The raw data were normalized using the robust multichip analysis (RMA) approach [70]. Linear models were used to assess the differential expression, while the empirical Bayes method was used to moderate the standard errors [71]. Differentially expressed genes were ranked by P values and fold changes. Genes with a cutoff threshold P value of 0.05 and LFC of ≥1.00 or ≤−1.00 were considered to be differentially expressed. To generate diagrams of the metabolic pathways and biological processes that were regulated, we used the Open Source MapMan 3.5.0 BETA program [24]. The gene ontology system in the MapMan program was used for the identification of the processes, pathways and gene families whose expression were significantly altered. Details of our microarray experiments and the MIAME-compliant microarray data have been deposited in the Gene Expression Omnibus database, the National Center of Biotechnology Information (Accession Number GSE33004).
Quantitative Reverse Transcription PCR (qRT-PCR) Assays
Expression of seven genes was confirmed using qRT-PCR and the same set of RNA that was used for the microarray. Primers were designed using the PrimerQuestSM program (Integrated DNA Technologies, Inc). All of the qRT-PCR reactions were performed using total volumes of 25 µl in an ABI PRISM 7900 sequence detection system (Applied Biosystems, Foster City, CA) with the QuantiTect SYBR Green RT-PCR Kit (Qiagen, Valencia, CA) and ∼50 ng of total RNA, 10 nM PCR primers, 0.25 µl RT mix (Qiagen, Valencia, CA) and 12.5 µl QuantiTect SYBR Green RT-PCR Master Mix (Qiagen, Valencia, CA). The PCR conditions were 30 min of reverse transcription at 50°C followed by 15 min of predenaturation at 95°C and 40 cycles of 15 s of denaturation at 94°C, 45 s of annealing at 55°C, and 30 s of extension at 72°C. The 18S rRNA gene was used as an endogenous control. At the end of the cycling phase, a dissociation curve was produced to ensure the specificity of the amplification. The relative expression ratios for each target gene were calculated using the 2–ΔΔCt method according to Livak and Schmittgen [72]. Details of the primers that were used and the genes that were tested are listed in Table 7.
Microscopy Analysis
Healthy and HLB affected round to triangular shaped young stems and roots showing primary and secondary growth were prepared for light and transmission electron microscopy using the following method. Samples were placed in 3% glutaraldehyde in 0.1 M potassium phosphate buffer, pH 7.2 for 4 hr at room temperature followed by 3 washes in the same buffer. Post-fixation in 2% osmium tetroxide in the same buffer was carried out for 4 hr at room temperature, then washed in two buffer changes before dehydration in acetone at 10% for 10 min each step. The samples were then infiltrated and embedded in Spurr’s resin [73]. One micrometer cross sections were made on an LKB Huxley Ultramicrotome (LKB Instruments Inc., Rockville, MD, USA) and stained with methylene blue/azure A, counter-stained with basic fuchsin [74]. These were photographed using a Leitz Labor-Lux S compound microscope (Leitz, Wetzlar, Germany) equipped with a Canon Power Shot S31 S digital camera. The same blocks were then thin sectioned with the same microtome, mounted on 200 mesh Formvar coated Cu grids followed by staining with 2% aq. uranyl acetate and post staining with Reynolds lead citrate [75]. The grids were examined with a Morgagni 268 transmission electron microscope (FEI, The Netherlands) equipped with an AMT digital camera (Danvers, MA, USA).
Supporting Information
Cellular pathways that are regulated by Ca . L. asiaticus infection in the stems and roots of Valencia sweet orange ( Citrus sinensis ). A = stem and B = root. Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red.
(TIF)
Regulatory pathways that are altered by Ca . L. asiaticus infection in the stems and roots of Valencia sweet orange ( Citrus sinensis ). A = stem and B = root. Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red.
(TIF)
Starch and sugar metabolic pathway genes that are regulated by Ca . L. asiaticus infection in the stems of Valencia sweet orange ( Citrus sinensis ). Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red. Abbreviations/definitions: ADP-glucose pyrophosphorylase large subunit 3 (APL3), granule-bound starch synthase (GBSS), acid invertase (ACI), vacuolar invertase (VAI), alpha-amylase (AMY), beta-amylase (BMY), and sugar transporter 1 (SUT1).
(TIF)
Regulation of transcription factor-encoding genes by Ca . L. asiaticus infection in the stems and roots of Valencia sweet orange ( Citrus sinensis ). A = stem and B = root. Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red. Abbreviations/definitions: ABI3/VP1, ABI3/VP1-related B3-domain-containing TF family; AP2/EREBP, APETALA2/ethylene-responsive element binding protein family; ARF, auxin response factor; bZIP, basic leucine zipper motif; bHLH, basic helix-loop-helix family; C2C2-CO-like, CONSTANS-like zinc finger family; C2C2-Dof, C2C2(Zn) Dof family; C2C2-YABBY, C2C2(Zn) YABBY family; C2C2-GATA, C2C2(Zn) GATA family; C2H2, C2H2 zinc finger family; C3H, C3H zinc finger family; CCAAT-DR1, CCAAT box binding factor DR1; ORPHAN, Orphan family; NAC, NAC domain; MYB-related, MYB-related family; MYB, MYB domain; MADS, MADS box domain; HB, homeobox TF family; HSF, heat shock TF family; GRF, GRF family; G2-like, G2-like family GARP; GRAS, GRAS family; E2F-DP, E2F/DP family; EIL, EIN3-like; CCAAT-HAP2, CCAAT box binding factor HAP2; CCP, CPP(Zn), CPP1-related family; SBP, SBP family; TCP, TCP domain TF; Global, Global TF group; High mobility, high mobility group (HMG) family; Trihelix, triple helix family; TUB, Tubby (TUB) homolog TF; Histone DAase, histone deacetylase; Histone ATse, histone acetyltransferase; WRKY, WRKY domain family; AS2, lateral organ boundary gene family; JUMONJI, JUMONJI class TF; AT-rich, AT-rich interaction domain-containing family; AtSR, AtSR family; LUG, LEUNIG (LUG) domain family; Methyl BD, methyl binding domain proteins; Aux/, Aux/family; B3, B3 DNA binding domain TF; NPR1, NPR1 family; NIN-like, NIN-like bZIP-related family; Bromodomain, bromodomain proteins; BZR, Brassinazole resistant TF family; Nucleosome assembly, nucleosome/chromatin assembly factor group; Chromatin Remodeling, chromatin remodeling factors; PHD finger, PHD finger family; PHOR1, photoperiod-responsive 1; Dicer-like, dicer; DNA MT, DNA methyltransferase; Polycomb, Polycomb group (PcG); Pseudo ARR, Pseudo ARR; ELF, ELF3 TF; FHA, Forkhead-associated (FHA) domain TF family; PWWP domain, PWWP domain protein family; SET-domain, SET-domain transcriptional regulator family; GeBP, GeBP-like family; General, general transcription; Silencing, silencing group; SNF7, SNF7 family; TAZ, Transcriptional co-activator with PDZ binding motif; CCHC, Zn-finger (CCHC).
(TIF)
Regulation of secondary metabolic pathway genes by Ca . L. asiaticus infection in the stems and roots of Valencia sweet orange ( Citrus sinensis ). A = stem and B = root. Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red.
(TIF)
Regulation of phenylpropanoid pathway genes by Ca . L. asiaticus infection in the stems of Valencia sweet orange ( Citrus sinensis ). Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red. There was no significantly up-regulated phenylpropanoid pathway genes observed in the roots.
(TIF)
Funding Statement
This work was supported by the Citrus Research and Development Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bové JM (2006) Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Path: 7–37.
- 2.Ogata T, Kobori Y, Kawabe K, Yonemoto H, Ohto Y, et al. (2008) The effects of HLB-infection on respiration and development of roots of feroniella rootstock (Feroniella oblata) which showed resistance to HLB bacterium. Gottwald TR, Graham JH (Eds) Proceedings of the 1st International Research Conference on Huanglongbing Orlando, FL, USA. (pp. 184).
- 3. Johnson E, Bright DB, Graham JH (2012) Early root infection and damage in citrus huanglongbing disease development. Phytopathology 102: S4.59. [Google Scholar]
- 4. Kim JS, Sagaram US, Burns JK, Li JL, Wang N (2009) Response of sweet orange (Citrus sinensis) to ‘Candidatus Liberibacter asiaticus’ infection: microscopy and microarray analyses. Phytopathology 99: 50–57. [DOI] [PubMed] [Google Scholar]
- 5. Achor DS, Etxeberria E, Wang N, Folimonova SY, Chung KR, et al. (2010) Sequence of anatomical symptom observations in citrus affected with Huanglongbing disease. Plant Pathol J 9: 56–64. [Google Scholar]
- 6. Etxeberria E, Gonzalez P, Achor D, Albrigo G (2009) Anatomical distribution of abnormally high levels of starch in HLB-affected Valencia orange trees. Physiol Mol Plant Pathol 74: 76–83. [Google Scholar]
- 7. Fan J, Chen C, Brlansky RH, Gmitter FG, Li ZG (2010) Changes in carbohydrate metabolism in Citrus sinensis infected with ‘Candidatus Liberibacter asiaticus’. Plant Pathol 59: 1037–1043. [Google Scholar]
- 8. Folimonova SY, Achor DS (2010) Early events of citrus greening (Huanglongbing) disease development at the ultrastructural level. Phytopathology 100: 949–958. [DOI] [PubMed] [Google Scholar]
- 9. Baldwin E, Plotto A, Manthey J, McCollum G, Bai J, et al. (2010) Effect of liberibacter infection (huanglongbing disease) of citrus on orange fruit physiology and fruit/fruit juice quality: chemical and physical analyses. J Agric Food Chem 58: 1247–1262. [DOI] [PubMed] [Google Scholar]
- 10. Dagulo L, Danyluk MD, Spann TM, Valim MF, Goodrich-Schneider R, et al. (2010) Chemical characterization of orange juice from trees infected with citrus greening (Huanglongbing). J Food Sci 75: C199–207. [DOI] [PubMed] [Google Scholar]
- 11. Rosales R, Burns JK (2011) Phytohormone changes and carbohydrate status in sweet orange fruit from Huanglongbing-infected trees. J Plant Growth Regul 30: 312–321. [Google Scholar]
- 12. Tatineni S, Sagaram US, Gowda S, Robertson CJ, Dawson WO, et al. (2008) In planta distribution of ‘Candidatus Liberibacter asiaticus’ as revealed by polymerase chain reaction (PCR) and real-time PCR. Phytopathology 98: 592–599. [DOI] [PubMed] [Google Scholar]
- 13. Albrecht U, Bowman K (2008) Gene expression in Citrus sinensis (L.) Osbeck following infection with the bacterial pathogen Candidatus Liberibacter asiaticus causing Huanglongbing in Florida. Plant Sci 175: 291–306. [Google Scholar]
- 14. Fan J, Chen C, Yu Q, Brlansky RH, Li ZG, et al. (2011) Comparative iTRAQ proteome and transcriptome analyses of sweet orange infected by “Candidatus Liberibacter asiaticus”. Physiol Plant 143: 235–245. [DOI] [PubMed] [Google Scholar]
- 15. Martinelli F, Uratsu SL, Albrecht U, Reagan RL, Phu ML, et al. (2012) Transcriptome profiling of citrus fruit response to huanglongbing disease. PLoS One 7: e38039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng ZL, Zhao Y (2013) Transcriptome comparison and gene coexpression network analysis provide a systems view of citrus response to ‘Candidatus Liberibacter asiaticus’ infection. BMC Genomics 14: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Albrecht U, Bowman KD (2012) Transcriptional response of susceptible and tolerant citrus to infection with Candidatus Liberibacter asiaticus. Plant Sci 185–186: 118–130. [DOI] [PubMed] [Google Scholar]
- 18. Liao HL, Burns JK (2012) Gene expression in Citrus sinensis fruit tissues harvested from huanglongbing-infected trees: comparison with girdled fruit. J Exp Bot 63: 3307–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mafra V, Martins PK, Francisco CS, Ribeiro-Alves M, Freitas-Astúa J, et al. (2013) Candidatus Liberibacter americanus induces significant reprogramming of the transcriptome of the susceptible citrus genotype. BMC Genomics 14: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kogenaru S, Qing Y, Guo Y, Wang N (2012) RNA-seq and microarray complement each other in transcriptome profiling. BMC Genomics 13: 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin JA, Wang Z (2011) Next-generation transcriptome assembly. Nat Rev Genet 12: 671–682. [DOI] [PubMed] [Google Scholar]
- 22. Zhou X, Ren L, Meng Q, Li Y, Yu Y, et al. (2010) The next-generation sequencing technology and application. Protein Cell 1: 520–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nwugo CC, Lin H, Duan Y, Civerolo EL (2013) The effect of ‘Candidatus Liberibacter asiaticus’ infection on the proteomic profiles and nutritional status of pre-symptomatic and symptomatic grapefruit (Citrus paradisi) plants. BMC Plant Biol 13: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, et al. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939. [DOI] [PubMed] [Google Scholar]
- 25. Stark DM, Timmerman KP, Barry GF, Preiss J, Kishore GM (1992) Regulation of the amount of starch in plant tissues by ADP glucose pyrophosphorylase. Science 258: 287–292. [DOI] [PubMed] [Google Scholar]
- 26. Seifert GJ, Blaukopf C (2010) Irritable walls: the plant extracellular matrix and signaling. Plant Physiol 153: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Somerville C (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78. [DOI] [PubMed] [Google Scholar]
- 28. Vogel J (2008) Unique aspects of the grass cell wall. Curr Opin Plant Biol 11: 301–307. [DOI] [PubMed] [Google Scholar]
- 29. Keegstra K, Walton J (2006) Plant science. Beta-glucans–brewer’s bane, dietician’s delight. Science 311: 1872–1873. [DOI] [PubMed] [Google Scholar]
- 30. Li M, Xiong G, Li R, Cui J, Tang D, et al. (2009) Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J 60: 1055–1069. [DOI] [PubMed] [Google Scholar]
- 31. Rösti J, Barton CJ, Albrecht S, Dupree P, Pauly M, et al. (2007) UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana . Plant Cell 19: 1565–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lamport DT, Kieliszewski MJ, Chen Y, Cannon MC (2011) Role of the extensin superfamily in primary cell wall architecture. Plant Physiol 156: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Darley CP, Forrester AM, McQueen-Mason SJ (2001) The molecular basis of plant cell wall extension. Plant Mol Biol 47: 179–195. [PubMed] [Google Scholar]
- 34. Campbell P, Braam J (1999) Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci 4: 361–366. [DOI] [PubMed] [Google Scholar]
- 35. Gray-Mitsumune M, Mellerowicz EJ, Abe H, Schrader J, Winzéll A, et al. (2004) Expansins abundant in secondary xylem belong to subgroup A of the alpha-expansin gene family. Plant Physiol 135: 1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gray-Mitsumune M, Abe H, Takahashi J, Sundberg B, Mellerowicz EJ (2004) Liquid phase fluorescence in situ RT-PCR analysis for gene expression analysis in woody stems. Plant Biol (Stuttg) 6: 47–54. [DOI] [PubMed] [Google Scholar]
- 37. Whitham S, McCormick S, Baker B (1996) The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proc Natl Acad Sci U S A 93: 8776–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Song HK, Suh SW (1998) Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator. J Mol Biol 275: 347–363. [DOI] [PubMed] [Google Scholar]
- 39. Ryan CA, Pearce G (2003) Systemins: a functionally defined family of peptide signals that regulate defensive genes in Solanaceae species. Proc Natl Acad Sci U S A 100 Suppl 214577–14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verica JA, Chae L, Tong H, Ingmire P, He ZH (2003) Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis. Plant Physiol 133: 1732–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shuai B, Reynaga-Peña CG, Springer PS (2002) The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol 129: 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Husbands A, Bell EM, Shuai B, Smith HM, Springer PS (2007) LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res 35: 6663–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Feng C, Andreasson E, Maslak A, Mock HP, Mattsson O, et al. (2004) Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci 167: 1099–1107. [Google Scholar]
- 44. Lai Z, Vinod K, Zheng Z, Fan B, Chen Z (2008) Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takatsuji H, Nakamura N, Katsumoto Y (1994) A new family of zinc finger proteins in petunia: structure, DNA sequence recognition, and floral organ-specific expression. Plant Cell 6: 947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Teakle GR, Manfield IW, Graham JF, Gilmartin PM (2002) Arabidopsis thaliana GATA factors: organisation, expression and DNA-binding characteristics. Plant Mol Biol 50: 43–57. [DOI] [PubMed] [Google Scholar]
- 47. Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, et al. (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120. [DOI] [PubMed] [Google Scholar]
- 48. Yanagisawa S, Sheen J (1998) Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lijavetzky D, Carbonero P, Vicente-Carbajosa J (2003) Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kovi MR, Zhang Y, Yu S, Yang G, Yan W, et al. (2011) Candidacy of a chitin-inducible gibberellin-responsive gene for a major locus affecting plant height in rice that is closely linked to Green Revolution gene sd1 . Theor Appl Genet 123: 705–714. [DOI] [PubMed] [Google Scholar]
- 51. Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, et al. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433. [DOI] [PubMed] [Google Scholar]
- 52. Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Duan Y, Zhou L, Hall DG, Li W, Doddapaneni H, et al. (2009) Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol Plant Microbe Interact 22: 1011–1020. [DOI] [PubMed] [Google Scholar]
- 54. Bürkle L, Cedzich A, Döpke C, Stransky H, Okumoto S, et al. (2003) Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J 34: 13–26. [DOI] [PubMed] [Google Scholar]
- 55. Wang WH, Köhler B, Cao FQ, Liu GW, Gong YY, et al. (2012) Rice DUR3 mediates high-affinity urea transport and plays an effective role in improvement of urea acquisition and utilization when expressed in Arabidopsis. New Phytol 193: 432–444. [DOI] [PubMed] [Google Scholar]
- 56. Schuster B, Rétey J (1995) The mechanism of action of phenylalanine ammonia-lyase: the role of prosthetic dehydroalanine. Proc Natl Acad Sci U S A 92: 8433–8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fan J, Chen C, Yu Q, Khalaf A, Achor DS, et al. (2012) Comparative transcriptional and anatomical analyses of tolerant rough lemon and susceptible sweet orange in response to ‘Candidatus Liberibacter asiaticus’ infection. Mol Plant Microbe Interact 25: 1396–1407. [DOI] [PubMed] [Google Scholar]
- 58.Wang N, Trivedi P (2013) Citrus Huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology. [DOI] [PubMed]
- 59. Scheidig A, Fröhlich A, Schulze S, Lloyd JR, Kossmann J (2002) Downregulation of a chloroplast-targeted beta-amylase leads to a starch-excess phenotype in leaves. Plant J 30: 581–591. [DOI] [PubMed] [Google Scholar]
- 60. DeYoung BJ, Innes RW (2006) Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol 7: 1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gao H, Narayanan NN, Ellison L, Bhattacharyya MK (2005) Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol Plant Microbe Interact 18: 1035–1045. [DOI] [PubMed] [Google Scholar]
- 62. Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, et al. (1995) Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant Microbe Interact 8: 863–870. [DOI] [PubMed] [Google Scholar]
- 63. Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic Acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206. [DOI] [PubMed] [Google Scholar]
- 64. Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, et al. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae . Plant Cell 15: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nomura K, Melotto M, He SY (2005) Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol 8: 361–368. [DOI] [PubMed] [Google Scholar]
- 66. Alleva K, Chara O, Amodeo G (2012) Aquaporins: another piece in the osmotic puzzle. FEBS Lett 586: 2991–2999. [DOI] [PubMed] [Google Scholar]
- 67. Jang JY, Rhee JY, Chung GC, Kang H (2012) Aquaporin as a membrane transporter of hydrogen peroxide in plant response to stresses. Plant Signal Behav 7: 1180–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yool AJ, Campbell EM (2012) Structure, function and translational relevance of aquaporin dual water and ion channels. Mol Aspects Med 33: 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Molina-Hidalgo FJ, Franco AR, Villatoro C, Medina-Puche L, Mercado JA, et al. (2013) The strawberry (Fragariaxananassa) fruit-specific rhamnogalacturonate lyase 1 (FaRGLyase1) gene encodes an enzyme involved in the degradation of cell-wall middle lamellae. J Exp Bot 64: 1471–1483. [DOI] [PubMed] [Google Scholar]
- 70. Bolstad BM, Irizarry RA, Åstrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- 71. Smyth GK, Michaud J, Scott HS (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075. [DOI] [PubMed] [Google Scholar]
- 72. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 73. Spurr AR (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43. [DOI] [PubMed] [Google Scholar]
- 74.Schneider H (1981) Plant anatomy and general botany. In: Clark G, editor. Staining procedures for biological stain commission. 4th ed. ed. Baltimore: Wms. and Wilkins. 315–373.
- 75. Reynolds ES (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17: 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cellular pathways that are regulated by Ca . L. asiaticus infection in the stems and roots of Valencia sweet orange ( Citrus sinensis ). A = stem and B = root. Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red.
(TIF)
Regulatory pathways that are altered by Ca . L. asiaticus infection in the stems and roots of Valencia sweet orange ( Citrus sinensis ). A = stem and B = root. Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red.
(TIF)
Starch and sugar metabolic pathway genes that are regulated by Ca . L. asiaticus infection in the stems of Valencia sweet orange ( Citrus sinensis ). Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red. Abbreviations/definitions: ADP-glucose pyrophosphorylase large subunit 3 (APL3), granule-bound starch synthase (GBSS), acid invertase (ACI), vacuolar invertase (VAI), alpha-amylase (AMY), beta-amylase (BMY), and sugar transporter 1 (SUT1).
(TIF)
Regulation of transcription factor-encoding genes by Ca . L. asiaticus infection in the stems and roots of Valencia sweet orange ( Citrus sinensis ). A = stem and B = root. Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red. Abbreviations/definitions: ABI3/VP1, ABI3/VP1-related B3-domain-containing TF family; AP2/EREBP, APETALA2/ethylene-responsive element binding protein family; ARF, auxin response factor; bZIP, basic leucine zipper motif; bHLH, basic helix-loop-helix family; C2C2-CO-like, CONSTANS-like zinc finger family; C2C2-Dof, C2C2(Zn) Dof family; C2C2-YABBY, C2C2(Zn) YABBY family; C2C2-GATA, C2C2(Zn) GATA family; C2H2, C2H2 zinc finger family; C3H, C3H zinc finger family; CCAAT-DR1, CCAAT box binding factor DR1; ORPHAN, Orphan family; NAC, NAC domain; MYB-related, MYB-related family; MYB, MYB domain; MADS, MADS box domain; HB, homeobox TF family; HSF, heat shock TF family; GRF, GRF family; G2-like, G2-like family GARP; GRAS, GRAS family; E2F-DP, E2F/DP family; EIL, EIN3-like; CCAAT-HAP2, CCAAT box binding factor HAP2; CCP, CPP(Zn), CPP1-related family; SBP, SBP family; TCP, TCP domain TF; Global, Global TF group; High mobility, high mobility group (HMG) family; Trihelix, triple helix family; TUB, Tubby (TUB) homolog TF; Histone DAase, histone deacetylase; Histone ATse, histone acetyltransferase; WRKY, WRKY domain family; AS2, lateral organ boundary gene family; JUMONJI, JUMONJI class TF; AT-rich, AT-rich interaction domain-containing family; AtSR, AtSR family; LUG, LEUNIG (LUG) domain family; Methyl BD, methyl binding domain proteins; Aux/, Aux/family; B3, B3 DNA binding domain TF; NPR1, NPR1 family; NIN-like, NIN-like bZIP-related family; Bromodomain, bromodomain proteins; BZR, Brassinazole resistant TF family; Nucleosome assembly, nucleosome/chromatin assembly factor group; Chromatin Remodeling, chromatin remodeling factors; PHD finger, PHD finger family; PHOR1, photoperiod-responsive 1; Dicer-like, dicer; DNA MT, DNA methyltransferase; Polycomb, Polycomb group (PcG); Pseudo ARR, Pseudo ARR; ELF, ELF3 TF; FHA, Forkhead-associated (FHA) domain TF family; PWWP domain, PWWP domain protein family; SET-domain, SET-domain transcriptional regulator family; GeBP, GeBP-like family; General, general transcription; Silencing, silencing group; SNF7, SNF7 family; TAZ, Transcriptional co-activator with PDZ binding motif; CCHC, Zn-finger (CCHC).
(TIF)
Regulation of secondary metabolic pathway genes by Ca . L. asiaticus infection in the stems and roots of Valencia sweet orange ( Citrus sinensis ). A = stem and B = root. Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red.
(TIF)
Regulation of phenylpropanoid pathway genes by Ca . L. asiaticus infection in the stems of Valencia sweet orange ( Citrus sinensis ). Genes that were significantly up-regulated following Ca. L. asiaticus infection are displayed in blue, and down-regulated genes are displayed in red. There was no significantly up-regulated phenylpropanoid pathway genes observed in the roots.
(TIF)