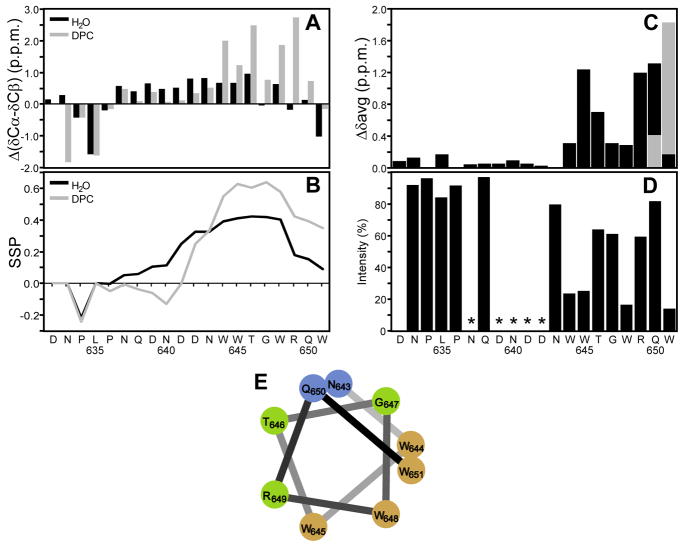
Figure 5.
Nuclear magnetic resonance of S-MPER (A) Chemical shift deviations from random coil shifts for δCα-δCβ. (B) Secondary structure propensity (SSP) scores for S-MPER in H2O (black) and in 200mM DPC micelles (gray) calculated using 13Cα, 13Cβ and 1Hα chemical shifts. Positive values represent helical propensity. (C) Average amide chemical shift differences, {[(Δδ1H)2 + 0.2(Δδ15N)2]/2}1/2, for S-MPER in H2O vs. 200mM DPC micelles. The alternative chemical shift difference for the ambiguous assignment of the amide correlations of Q19/W20 is shown in gray. (D) Effect of a paramagnetic agent on S-MPER in 200mM DPC micelles in D2O for the correlation of the last protonated carbon of each amino acid in the aliphatic 1H,13C HSQC spectrum (except that the Cγ/Hγ and H/C3ε correlations were used for Pro and Trp). The ratio of integrated crosspeak intensities with and without 9mM 5-doxyl stearate is plotted along with the amino acid sequence. Residues where it was not possible to get reliable intensity ratios because of overlapping crosspeaks are marked with *. (E) α-Helical wheel representation of the C-terminal residues (12–20) of S-MPER color coded by % intensity loss on addition of 9 mM 5-doxylstearate; >50% brown, 35–50% green, <35% blue.