Summary
Sporulating Bacillus subtilis cells assemble a transenvelope secretion complex that connects the mother cell and developing spore. The forespore protein SpoIIQ and the mother-cell protein SpoIIIAH interact across the double membrane septum and are thought to assemble into a channel that serves as the basement layer of this specialized secretion system. SpoIIQ is absolutely required to recruit SpoIIIAH to the sporulation septum on the mother-cell side, however the mechanism by which SpoIIQ is localized has been unclear. Here, we show that SpoIIQ localization requires its partner protein SpoIIIAH and degradation of the septal peptidoglycan (PG) by the two cell wall hydrolases SpoIID and SpoIIP. Our data suggest that PG degradation enables a second mother-cell-produced protein to interact with SpoIIQ. Cells in which both mother-cell anchoring mechanisms have been disabled have a synergistic sporulation defect suggesting that both localization factors function in the secretion complex. Finally, we show that septal PG degradation is critical for the assembly of an active complex. Altogether, these results suggest that the specialized secretion system that links the mother cell and forespore has a complexity approaching those found in Gram-negative bacteria and reveal that the sporulating cell must overcome similar challenges in assembling a transenvelope complex.
Keywords: LytM, protein localization, sporulation, cell wall hydrolase, specialized secretion
Introduction
During spore formation in Bacillus subtilis, the mother cell and forespore follow distinct programs of developmental gene expression that are linked to each other through cell-cell signaling pathways (Errington, 2003, Hilbert & Piggot, 2004, Higgins & Dworkin, 2012). In addition to these traditional modes of communication, the mother cell and forespore assemble a transenvelope secretion complex that spans the double membrane that separates them (Meisner et al., 2008, Camp & Losick, 2008, Doan et al., 2009, Camp & Losick, 2009, Meisner et al., 2012, Levdikov et al., 2012). This complex is required to maintain forespore physiology and for forespore gene expression at late stages in development (Doan et al., 2009, Camp & Losick, 2009). Little is known about this complex that appears to be assembled from components normally found in distinct Gram-negative secretion systems (Camp & Losick, 2008, Doan et al., 2009, Meisner et al., 2008). It is not known what this machine transports; what it looks like and how it is assembled; nor is it known whether a complete parts list for the complex has been identified. Here, we provide evidence for the involvement of an additional mother-cell protein in the assembly of the complex and show that, like Gram-negative secretion systems, degradation of peptidoglycan is important for the assembly of an active machine.
Upon the initiation of sporulation, the developing cell divides asymmetrically, generating two cells of unequal size and dissimilar fate (Higgins & Dworkin, 2012, Hilbert & Piggot, 2004, Errington, 2003). The smaller cell differentiates into the dormant spore and is called the forespore. The larger cell nurtures the developing spore and is referred to as the mother cell. The two cells follow distinct programs of gene expression that are linked to each other by cell-cell signaling pathways. At early stages of sporulation, forespore transcription under the control of σF is required to activate the mother-cell transcription factor σE. At late stages, the forespore transcription factor σG is required to activate mother-cell gene expression under σK control. Initially, the forespore and mother cell lie side by side separated by a double membrane septum. However, shortly after polar division, the first mother-cell transcription factor σE directs the synthesis of cell wall hydrolases that degrade the septal peptidoglycan and facilitate the migration of the mother-cell membranes around the forespore in a process called engulfment. Upon completion of engulfment, a membrane fission event releases the forespore into the mother-cell cytoplasm as a free protoplast surrounded by two membranes. At this stage, the mother packages the forespore in protective layers in preparation for dormancy. When the spore is fully mature, the mother cell lyses releasing it into the environment, where it stays dormant until nutrients trigger germination and outgrowth.
During the morphological process of engulfment, the mother cell and forespore assemble a transenvelope complex that spans the double membrane that separates them. Several lines of evidence suggest that the basement layer of this complex is composed of the mother-cell membrane protein SpoIIIAH and the forespore membrane protein SpoIIQ. First, the extracytoplasmic domains of these two proteins interact in the space between the two membranes (Blaylock et al., 2004, Doan et al., 2005). Second, SpoIIIAH shares weak sequence similarity with inner membrane ring proteins, YcsJ/EscJ and FliF, found in type III secretion systems and the flagellar basal body (Camp & Losick, 2008, Meisner et al., 2008); and recent co-crystal structures of the extracytoplasmic domains of SpoIIIAH and SpoIIQ revealed that SpoIIIAH is indeed a structural homolog of EscJ (Meisner et al., 2012, Levdikov et al., 2012). Finally, forespore-expressed biotin ligase can access and biotinylate the extracytoplasmic domain of SpoIIIAH in a SpoIIQ-dependent manner (Meisner et al., 2008). Collectively, these findings suggest that SpoIIIAH and SpoIIQ form a channel that connects the mother cell and forespore. Seven additional mother-cell proteins encoded in the spoIIIA operon (along with SpoIIIAH) are required to assemble an active secretion complex (Doan et al., 2009, Camp & Losick, 2009). Several of these proteins share sequence similarity to components of Type II/IV or Type III secretion systems found in Gram-negative bacteria (Camp & Losick, 2008, Meisner et al., 2008, Doan et al., 2009). In the absence of any of the SpoIIIA proteins or SpoIIQ, the forespore appears to collapse and lose its integrity (Doan et al., 2009). Moreover, consistent with a loss of integrity, gene expression in the forespore compartment ceases, suggesting that the forespore lose its metabolic potential (Camp & Losick, 2009). It is not known what this transenvelope complex transports, however, the presence of the secretion ATPase on the mother-cell side and the phenotypes of the forespore in the absence of the components of the machine suggest that transport occurs from mother cell to forespore and that the secreted factor(s) are necessary to maintain forespore metabolic potential and forespore gene expression (Camp & Losick, 2009, Doan et al., 2009).
Assembly of this apparatus appears to begin with the interaction of SpoIIIAH and SpoIIQ across the polar septum. SpoIIIAH localization to the septal membrane requires SpoIIQ in the forespore (Blaylock et al., 2004, Doan et al., 2005). Interestingly, SpoIIQ localizes to the septum on the forespore side in the absence of SpoIIIAH (Blaylock et al., 2004, Doan et al., 2005). The underlying mechanism by which SpoIIQ is anchored in the septal membrane is not known. However, Pogliano and co-workers have shown that in cells lacking the mother-cell transcription factor σE, SpoIIQ becomes mislocalized and is found in all membranes of the forespore (Blaylock et al., 2004, Rubio & Pogliano, 2004) (Fig. 1). This observation indicates that one (or several) mother-cell proteins under σE control are required to localize SpoIIQ and suggests that these localization factors could function in the assembly of the secretion complex. To gain a deeper understanding of this transenvelope machine, we set out to identify the localization determinants of SpoIIQ.
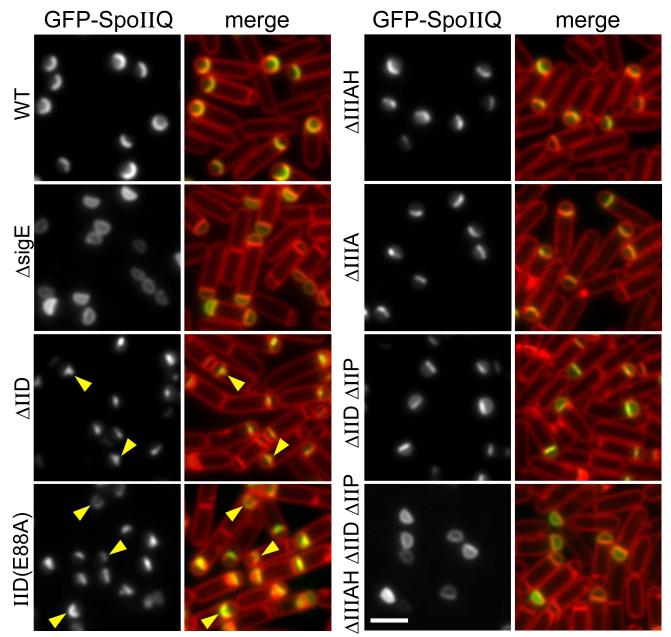
Figure 1. Localization of GFP-SpoIIQ to the forespore septal membrane requires SpoIID, SpoIIP and SpoIIIAH.
Representative images of GFP-SpoIIQ localization in sporulating cells at hour 2 of sporulation. Images are from wild-type (BKM1897), ΔsigE (BKM1930), ΔspoIIIAH (BKM1905), ΔspoIID (BKM1898), spoIID(E88A) (BKM1902), ΔspoIIIA operon (BCR24), a ΔspoIID ΔspoIIP double mutant (BKM1928) and a ΔspoIID ΔspoIIP ΔspoIIIAH triple mutant (BKM1929). Images are GFP-SpoIIQ (left) and merge of GFP-SpoIIQ with membranes stained with TMA-DPH. Localization of GFP-SpoIIQ to the bulged septal membrane is highlighted (yellow carets). Scale bar is 2 μm.
Here, we report that septal localization of SpoIIQ requires the σE-controlled proteins SpoIIIAH, SpoIID, and SpoIIP. Unlike SpoIIIAH that helps anchor SpoIIQ through direct protein-protein interaction, we show that SpoIID and SpoIIP act indirectly. Our data indicate that the catalytic activities of SpoIID and SpoIIP are necessary for SpoIIQ localization and suggest that septal PG degradation by these cell wall hydrolases allows an additional mother-cell protein under σE control to interact with and participate in anchoring SpoIIQ in the septal membrane. Mutational analysis supports the idea that this interaction occurs in the LytM groove of SpoIIQ. Cells in which both anchoring mechanisms have been disabled are severely impaired in sporulation, suggesting that both localization factors function in the secretion complex that maintains forespore integrity and late forespore gene expression. Finally, we show that peptidoglycan degradation in the intermembrane space is critical for the assembly of an active secretion complex. These results suggest that the secretion apparatus assembled during sporulation may have a complexity akin to those found in Gram-negative bacteria and that B. subtilis has come up with similar solutions to the challenges of assembling a transenvelope machine.
Results
SpoIID, SpoIIP and SpoIIIAH are required for proper localization of SpoIIQ
In the course of our analysis of the cell wall hydrolases SpoIID and SpoIIP (Morlot et al., 2010), we noticed that the forespore membrane protein SpoIIQ frequently localized within the septal bulge that was generated in strains harboring point mutants in SpoIID (Fig. 1). Under these conditions, SpoIID and SpoIIP are also present at or near these bulges on the mother cell side and fail to migrate around the forespore (Gutierrez et al., 2010). Accordingly, we wondered whether SpoIID and/or SpoIIP participate in localizing SpoIIQ in the septal membrane. To investigate this, we monitored the localization of a fully functional GFP-SpoIIQ fusion during sporulation in wild-type and in cells lacking SpoIID, SpoIIP, or both. In the absence of either SpoIID or SpoIIP the septal membrane bulges into the mother cell cytoplasm and GFP-SpoIIQ was frequently localized within the bulge (Rubio & Pogliano, 2004) (Fig. 1 and data not shown). In the absence of both proteins, degradation of the septal peptidoglycan is blocked and the asymmetric septum remains flat (Pogliano et al., 1999, Abanes-De Mello et al., 2002, Eichenberger et al., 2001) (Fig. 1). Under these conditions, GFP-SpoIIQ principally localized to the septal membrane with a small amount of protein present throughout the peripheral forespore membranes (Fig. 1). This pattern of SpoIIQ localization was qualitatively similar to what has been observed previously in cells lacking SpoIIIAH (Blaylock et al., 2004, Rubio & Pogliano, 2004) (Fig. 1). Interestingly, in a subset of sporulating cells lacking both SpoIID and SpoIIP (8%, n>600), GFP-SpoIIQ was completely mislocalized and was uniformly distributed throughout the forespore membranes (Fig. S1).
It has been reported previously that SpoIIIAH is less enriched at the sporulation septum in mutants blocked in septal PG thinning (Blaylock et al., 2004). Accordingly, the partial mislocalization of GFP-SpoIIQ in cells lacking SpoIID and SpoIIP could be due to reduced SpoIIIAH at the septum. Alternatively, the three σE-controlled genes (spoIIIAH, spoIID, spoIIP) could be responsible for localizing SpoIIQ in the septal membrane. To test this, we examined GFP-SpoIIQ localization during spore formation in a ΔspoIIP, ΔspoIID, ΔspoIIIAH triple mutant. Strikingly, in most cells, GFP-SpoIIQ was localized uniformly throughout the forespore membrane, closely mimicking the localization observed in a sigE null mutant (Fig. 1). In a subset of the triple mutant cells (16%, n>600), there was a small degree of enrichment of GFP-SpoIIQ in the septal membrane (Fig. S1), suggesting that additional σE-controlled genes participate in SpoIIQ localization. Nonetheless, our data indicate that SpoIIIAH, SpoIID, and SpoIIP play critical roles in localizing SpoIIQ. The requirement for these proteins in SpoIIQ localization was independently discovered by K. Pogliano and co-workers (Fredlund et al., 2013).
SpoIIQ localizes in the wake of SpoIID and SpoIIP during the morphological process of engulfment
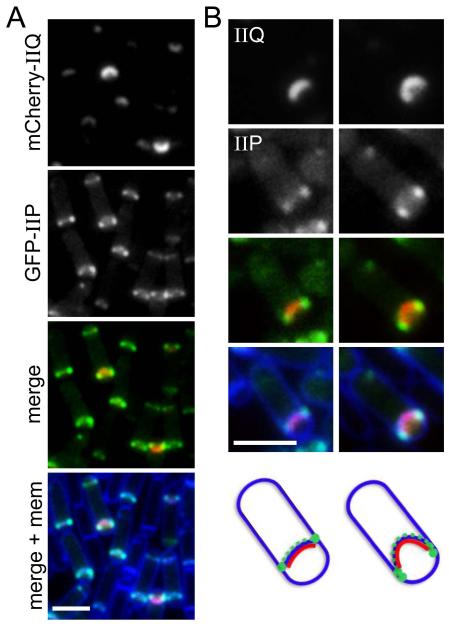
SpoIID and SpoIIP have been reported previously to localize at the leading edge of the engulfing septal membrane where they degrade the cell wall peptidoglycan and are thought to pull or guide the mother-cell membranes around the forespore (Abanes-De Mello et al., 2002, Chastanet & Losick, 2007, Aung et al., 2007, Morlot et al., 2010, Gutierrez et al., 2010). By contrast, during engulfment, SpoIIQ localizes more uniformly along the septal membrane on the forespore side (Rubio & Pogliano, 2004) (Fig. 1). Accordingly, it seemed unlikely that SpoIID and SpoIIP anchor SpoIIQ through direct protein-protein interactions. However, to compare their localization in the same cells, we performed double-labeling experiments using a mCherry-SpoIIQ fusion and either GFP-SpoIID or GFP-SpoIIP. Strains harboring the two fluorescent fusions as the sole source of these proteins sporulated at levels similar to wild-type and had no significant defects in engulfment (data not shown). At hour 2 of sporulation when the mother-cell membranes were migrating around the forespore, mCherry-SpoIIQ localized along the septal membrane as a curved band, or in sporulating cells that had just initiated the engulfment process, as a focus at the center of the forespore septal membrane (Fig. 2A and SD1). By contrast, GFP-SpoIIP (and separately GFP-SpoIID) localized weakly along the septal membrane, presumably on the mother-cell side, and as distinct and bright foci at the leading edge of the engulfing septal membranes (Fig. 2 and S2B). In merged images, mCherry-SpoIIQ appeared to localize behind the foci of GFP-SpoIIP suggesting that SpoIIQ localizes in the wake of the cell wall degrading activities of SpoIID and SpoIIP. Similar results were obtained using a GFP-SpoIID fusion (Fig. S2A and S2B). These data favor the idea that SpoIID and SpoIIP do not physically interact with SpoIIQ and therefore only indirectly participate in localizing it. However, since some GFP-SpoIIP and GFP-SpoIID were present in the septal membrane behind the leading edge of the engulfing membrane, these experiments cannot rule out a direct protein-protein interaction in the intermembrane space.
Figure 2. mCherry-SpoIIQ localizes in the wake of GFP-SpoIIP.
Double labeling of mCherry-SpoIIQ and GFP-SpoIIP in sporulating cells (BCR188) imaged at hour 2 of sporulation. (A) Representative fields showing mCherry-SpoIIQ, GFP-SpoIIP, a merged image of the two fluorescent fusion proteins and a merged image including the membranes stained with TMA-DPH. (B) Larger images highlighting examples where mCherry-SpoIIQ and GFP-SpoIIP do not completely overlap. Schematic representations of mCherry-SpoIIQ (red), GFP-SpoIIP (green) and membranes (blue) are shown below. Scale bar is 2 μm. We note that mCherry-SpoIIQ localization is slightly delayed compared to GFP-SpoIIQ. We attribute this lag to the maturation times of the mCherry compared to GFP (Cormack et al., 1996, Shaner et al., 2005, Merzlyak et al., 2007). Fig. S2B contains additional representative examples of cells from the same experiment.
SpoIID and SpoIIP catalytic activities are required for SpoIIQ localization
The double-labeling experiment described above suggests that SpoIIQ localizes to the septal membrane in places where SpoIID and SpoIIP have thinned the septal PG. If this is true, then catalytic mutants of SpoIID and SpoIIP should be defective in localizing SpoIIQ. To test this, we generated a strain in which the only copy of SpoIID was the catalytic mutant SpoIIDE88A and the only copy of SpoIIP was SpoIIPE359A (Morlot et al., 2010, Shida et al., 2001). Sporulating cells harboring these two catalytic point mutants phenocopied a ΔspoIID, ΔspoIIP double mutant: engulfment was completely blocked and GFP-SpoIIQ localized to the septal membrane with a small amount of protein present throughout the peripheral forespore membranes (data not shown).
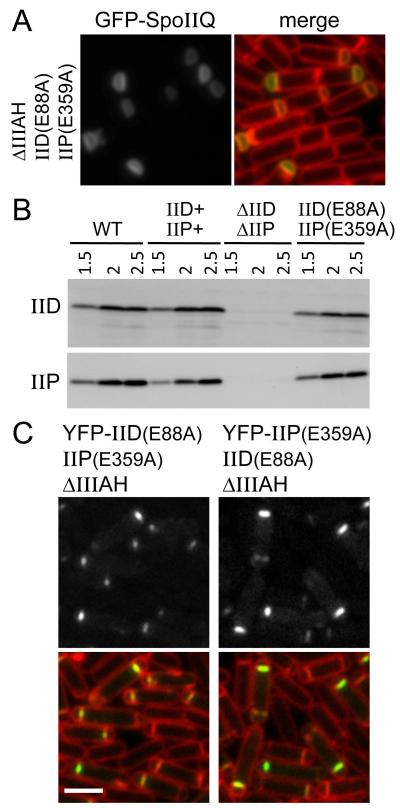
Next, we introduced the ΔspoIIIAH deletion into the strain harboring the catalytic point mutants and monitored GFP-SpoIIQ localization during sporulation. In most cells, GFP-SpoIIQ localized uniformly throughout the forespore membranes, with no obvious enrichment at the forespore septal membrane, mimicking the localization observed in ΔspoIID, ΔspoIIP, ΔspoIIIAH triple mutant and the sigE null (Fig. 3A and Fig. 1). Importantly, both catalytic mutants were expressed at levels comparable to their wild-type counterparts (Fig. 3B). Moreover, YFP fusions to both mutants localized to the asymmetric septum in match-controlled genetic backgrounds (Fig. 3C). Collectively, these data suggest that the catalytic activities of SpoIID and SpoIIP are required for the proper localization of SpoIIQ. Although we cannot exclude the possibility that the mutated residues in SpoIID and SpoIIP are required for interaction with SpoIIQ, in light of the double-labeling experiments described above, we favor the idea that the role of SpoIID and SpoIIP in SpoIIQ localization is in degrading the cell wall peptidoglycan that resides between the double membrane separating the mother cell and forespore.
Figure 3. The catalytic activities of SpoIID and SpoIIP are required for GFP-SpoIIQ localization to the forespore septal membrane.
(A) Representative images of GFP-SpoIIQ localization in the ΔspoIIIAH, SpoIIDE88A SpoIIPE359A mutant (strain BCR12) at hour 2 of sporulation. Images are GFP-SpoIIQ (left) and merged with TMA-DPH-stained membranes (right). Scale bar is 2 μm. (B) Immunoblot analysis showing that cells harboring spoIID(E88A) and spoIIP(E359A) produce wild-type levels of protein. Cell lysates of wild-type (BKM1897), ΔspoIIIAH spoIID+ spoIIP+ (BCR13), ΔspoIIIAH ΔspoIID ΔspoIIP (BKM1929) and ΔspoIIIAH spoIID(E88A) spoIIP(E359A) (BCR12) were collected at hour 1.5, 2, and 2.5 after the onset of sporulation and analyzed for SpoIID and SpoIIP protein level. (C) SpoIIDE88A and SpoIIPE359A localize to the polar septum. Representative images of YFP-SpoIIDE88A and YFP-SpoIIPE359A at hour 2 of sporulation in a ΔspoIIIAH spoIIP(E359A) background (BCR143) and ΔspoIIIAH spoIID(E88A) background (BCR121), respectively. Scale bar is 2 μm.
SpoIID and SpoIIP activities allow another σE-controlled protein to anchor SpoIIQ
How might the degradation of septal peptidoglycan by SpoIID and SpoIIP promote specific localization of SpoIIQ to the septal membrane? Since SpoIIQ contains a degenerate LytM domain that in some proteins confers PG binding activity (Uehara et al., 2009, Sabala et al., 2012), one possibility that we considered is that thinning of the septal PG by SpoIID and SpoIIP generates modifications to the cell wall to which SpoIIQ binds. However, extensive attempts by us, and others (Meisner & Moran, 2011), to uncover an interaction between SpoIIQ and PG (with or without treatment by SpoIID and SpoIIP) were unsuccessful (Fig. S3). Accordingly, we favor a second model in which an additional mother-cell anchoring protein interacts with SpoIIQ across the septal membrane. However, unlike SpoIIIAH, which can localize SpoIIQ in the presence of septal PG (Fig. 1; ΔspoIIP ΔspoIID), this additional protein requires thinning of the PG to interact with SpoIIQ across the intermembrane space.
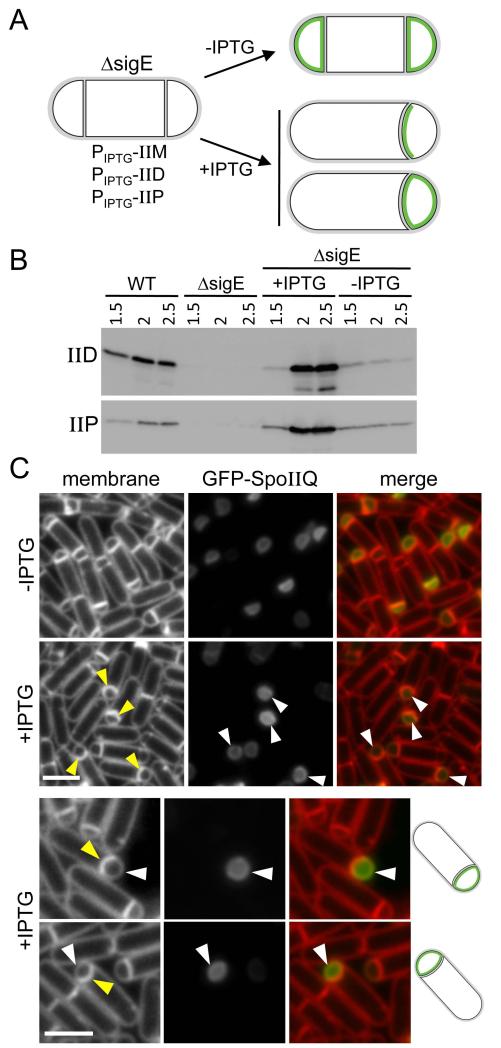
If such a protein exists, it is either produced under the control of σE or it is synthesized prior to σE activation. To distinguish between these two possibilities, we generated a sigE null mutant strain that expresses SpoIID, SpoIIP and their partner protein SpoIIM under IPTG-inducible control (Fig. 4A). SpoIIM is a polytopic membrane protein produced under σE control that is thought to form a complex with SpoIID and SpoIIP and help localize this cell wall degrading machine to the septal membrane (Eichenberger et al., 2001, Chastanet & Losick, 2007, Aung et al., 2007). We reasoned that if the second anchoring protein is produced independently of σE, then septal thinning by SpoIID and SpoIIP should be sufficient to localize GFP-SpoIIQ to the forespore membrane. If, on the other hand, this second protein is under σE control, then GFP-SpoIIQ should fail to localize even if the septal PG had been degraded (Fig. 4A).
Figure 4. An additional σE-controlled gene is required to localize GFP-SpoIIQ at the polar septum.
(A) Experimental rationale and potential outcomes for GFP-SpoIIQ localization (in green) when SpoIID, SpoIIP and SpoIIM are artificially produced in a ΔsigE mutant (strain BCR299). (B) Immunoblot analysis showing that SpoIID and SpoIIP are produced at wild-type levels upon addition of 1 mM IPTG to the culture medium 1.5 hours after the onset of sporulation. Cell lysates of strain BCR299 in the presence and absence of IPTG were analyzed for SpoIID and SpoIIP protein levels. As controls, a wild-type strain (BCR46) and a ΔsigE mutant (BKM1930) were included. (C) Representative images of cells (strain BCR299) at hour 2 of sporulation from cultures with and without IPTG addition. Engulfing septal membranes (yellow carets) and uniform GFP-SpoIIQ localization in the forespore membranes (white caret) are indicated. Images are TMA-DPH-stained membranes (left), GFP-SpoIIQ (middle) and a merge of the two (right). Scale bar is 2 μm. We note that in a subset of sporulating cells in which IPTG was added we observed a loss of GFP-SpoIIQ compartmentalization (see Fig. S5).
We induced sporulation in the sigE null mutant engineered to produce SpoIID, SpoIIP, and SpoIIM and analyzed GFP-SpoIIQ localization in the presence and absence of IPTG (Fig. 4A). In the absence of IPTG, the septal membranes remained flat in the vast majority of cells and, as expected, GFP-SpoIIQ localized uniformly in the forespore membranes (Fig. 4C). In a parallel culture, IPTG (1mM final) was added at hour 1.5 of sporulation at a time when most sporulating cells were undergoing asymmetric division. Thirty minutes later, when both SpoIID and SpoIIP were produced to levels similar to wild-type, as assayed by immunoblot (Fig. 4B), the polar septa began to curve (Fig. 4C, yellow carets) indicative of active thinning of the septal PG and the onset of engulfment. Importantly, in the sporulating cells that contained curved septa, GFP-SpoIIQ localized uniformly throughout the forespore membranes, with no specific localization at the septum (Fig. 4C, white carets). Collectively, these data support the idea that SpoIIIAH and a second protein under σE control interact with and localize SpoIIQ in the septal membrane.
Mutations in the LytM groove of SpoIIQ define a SpoIIIAH-independent interaction surface
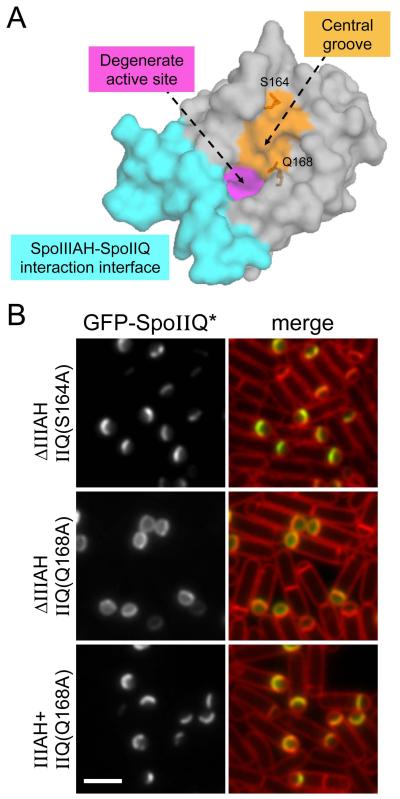
We reasoned that if a second protein interacts with SpoIIQ that it would bind SpoIIQ on a different surface from the one to which SpoIIIAH binds. Accordingly, we examined the recently published crystal structures of the SpoIIQ-SpoIIIAH complex (Meisner et al., 2012, Levdikov et al., 2012). One region of the SpoIIQ protein that is not occluded by SpoIIIAH includes the groove in the degenerate LytM domain (Fig. 5A). In active PG hydrolases that contain a LytM domain, the residues within the groove are required to interact with peptidoglycan (Sabala et al., 2012). Interestingly, these enzymes are produced in a latent form that is held inactive by their amino-terminal segments, which occlude the catalytic residues within the LytM groove (Odintsov et al., 2004). Thus, this groove can also engage in protein-protein interactions. Accordingly, we identified surface exposed residues in the LytM groove and mutated them individually to Alanine in the context of the GFP-SpoIIQ fusion (Fig. 5A and Fig. S4). We then tested these mutants for their ability to localize in a wild-type background and in cells lacking SpoIIIAH. If these mutants disrupt the interaction between SpoIIQ and a second σE-controlled protein then the mutant proteins should still localize to the septal membrane on their own but fail to localize in the absence of SpoIIIAH. The localization phenotypes of the various mutants could be broadly categorized into four classes: 1) mutants that had no impact on GFP-SpoIIQ localization (S164A, V166A and E156A); 2) mutants that were uniformly localized in the forespore membranes at early stages of sporulation but became cytoplasmic at later stages, presumably due to degradation (H202A, E206A and R208A); 3) mutants that were almost uniformly localized in the forespore membranes throughout engulfment (D123A and Q168A), and 4) mutants that produced a mixture of class 1 and 2 (S119A) (Fig. 5 and S4). Class 3 mutants had the expected phenotype and we present the data for one of these (SpoIIQQ168A) in Figure 5.
Figure 5. Point mutants in the LytM groove of SpoIIQ impair its localization to the forespore septal membrane.
(A) Surface representation of the SpoIIQ crystal structure highlighting the SpoIIIAH-SpoIIQ interaction interface (cyan), the degenerate active site (magenta) and the characteristic LytM groove (orange). Representative residues in the LytM groove (Q168 and S164) that were mutated are shown. (B) Images of GFP-SpoIIQ localization at hour 2 of sporulation. A representative mutant that localized properly (GFP-SpoIIQS164A) (BCR71) and one that failed to localize (GFP-SpoIIQQ168A)(BCR80). The loss of septal localization of GFP-SpoIIQQ168A was dependent on the spoIIIAH mutant. GFP-SpoIIQQ168A localized properly in an otherwise wild-type background (spoIIIAH+, BCR87). Scale bar is 2 μm.
In the presence of wild-type SpoIIIAH, the GFP-SpoIIQQ168A mutant (and the other class 3 mutant) retained its specific localization at the forespore septum (Fig. 5B and data not shown). However, in cells lacking SpoIIIAH, the GFP-SpoIIQQ168A mutant (and GFP-SpoIIQD123A) was almost completely mislocalized (Fig. 5B and S4). For comparison, we examined conservative and non-conservative point mutants (S164A, V166A and E156A) that also reside in the LytM groove (Fig 5A and Fig. S4). These mutants had no impact on localization of SpoIIQ in the presence or absence of SpoIIIAH. Altogether, these results identify residues within the LytM groove of SpoIIQ that are important for its localization and likely make up an interaction surface for a second mother-cell anchoring protein.
The second SpoIIQ anchoring protein helps maintain forespore integrity and σG activity
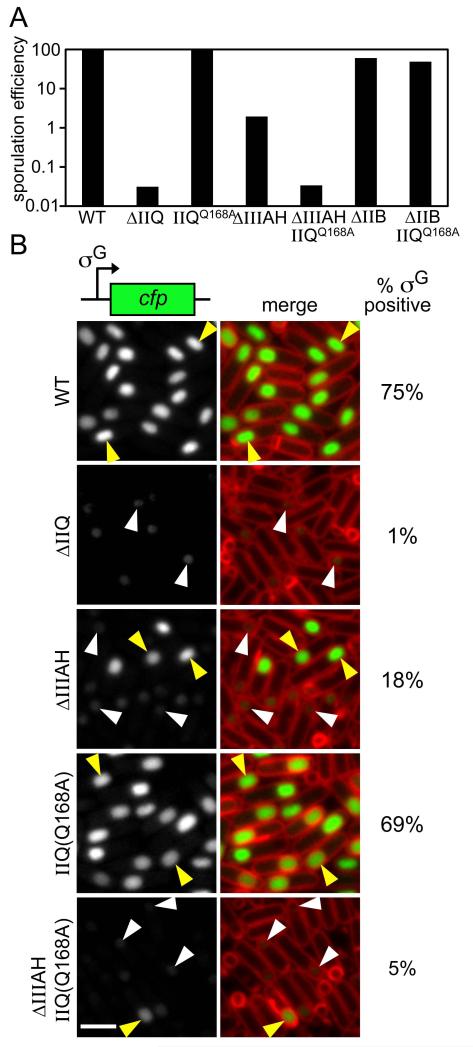
Our data are consistent with a model in which SpoIID and SpoIIP degrade septal PG to allow SpoIIQ to interact with a second σE-controlled protein. To investigate whether the second anchoring protein also functions in the secretion apparatus that maintains forespore physiology, we took advantage of the class 3 mutant (SpoIIQQ168A) that fails to localize in the absence of SpoIIIAH. Specifically, we examined the affect of the SpoIIQQ168A mutant on the production of heat-resistant spores and on its ability to activate the forespore transcription factor σG that requires an intact transenvelope complex (Doan et al., 2009, Camp & Losick, 2009). σG activity was assessed in single cells by fluorescence microscopy using a σG-responsive promoter (PsspB) fused to cfp (Doan et al., 2009).
Alone, the SpoIIQQ168A mutant had no discernable impact on the production of heat-resistant spores (Fig. 6A; Table S1). Moreover, it did not affect the activation of σG. At 3.5 hours after the onset of sporulation, 69% of the SpoIIQQ168A sporulating cells (n>600) contained strong fluorescent signal in the forespore (Fig. 6B, yellow caret). A similar percentage of sporulating cells (75%) had σG activity in wild-type (Fig. 6B). Since the SpoIIQQ168A mutant resulted in SpoIIQ mislocalization only in the absence of SpoIIIAH, we reasoned that its effect on sporulation would be more pronounced in a spoIIIAH null background. To this end, we examined the production of heat-resistant spores and σG activity of a ΔspoIIIAH, spoIIQQ168A double mutant. We observed a 60-fold reduction in the number of heat-resistant spores in the ΔspoIIIAH, spoIIQQ168A mutant compared to the ΔspoIIIAH mutant alone (Fig. 6A; Table S1). Importantly, the double mutant produced almost the same number of heat-resistant spores as cells lacking SpoIIQ (Fig. 6A; Table S1). This synthetic sporulation defect was specific to the spoIIIAH null, since spoIIQQ168A had no impact on the sporulation efficiency of a strain lacking a sporulation protein (SpoIIB) that is not involved in the secretion apparatus (Perez et al., 2000) (Fig. 6A). Importantly, we also observed a reduction in the number of cells with σG activity in the ΔspoIIIAH, spoIIQQ168A mutant relative to the ΔspoIIIAH null (Fig 6B and S6). At hour 3.5 of sporulation, only 5% (n>600) of the ΔspoIIIAH, spoIIQQ168A mutant cells contained strong σG-dependent CFP fluorescence in the forespore, compared to 18% (n>600) in the spoIIIAH null mutant (Fig. 6B, yellow carets). Finally, as reported previously (Doan et al., 2009), we observed that the forespores containing little or no CFP signal (i.e. reduced or no σG activity) were also smaller in appearance (Fig 6 and S6, white carets). This reduced size is indicative of a collapsed forespore (Doan et al., 2009), and was particularly pronounced in cells lacking SpoIIQ and in the ΔspoIIIAH, spoIIQQ168A double mutant. Altogether, these results suggest that the second anchoring protein also functions in the secretion apparatus and can partially compensate for the absence of SpoIIIAH (see Discussion).
Figure 6. The interaction mediated by the LytM groove in SpoIIQ is required for efficient sporulation and σG activity.
(A) Bar graph showing sporulation efficiency of wild-type (BCR163), ΔspoIIQ (BTD1541), spoIIQ(Q168A) (BCR152), ΔspoIIIAH (BCR161), spoIIQ(Q168A), ΔspoIIIAH double mutant (BCR153), ΔspoIIB (BCR162) and a spoIIQ(Q168A), ΔspoIIB double mutant (BCR196). (B) Representative images of strains harboring the σG-responsive fluorescent reporter PsspB-cfp at hour 3.5 of sporulation. From top to bottom: wild-type (BCR186), ΔspoIIQ (BCR151), ΔspoIIIAH (BCR193), spoIIQ(Q168A) (BCR185) and spoIIQ(Q168A) ΔspoIIIAH double mutant (BCR191). Forespores that display σG activity and are scored as positive (yellow carets) and those with weak activity and a collapsed forespore that are scored as negative (white carets) are highlighted (see Materials and Methods). The percentages of σG positive cells are indicated on the right. We note that σG-dependent gene expression appears higher in our assays than in those reported previously using lacZ fusions (Camp & Losick, 2008). We suspect this reflects the difference between single-cell- and population-based assays and the threshold used to score σG in an “on” vs. “off” state. Images are CFP (left) and merge with TMA-DPH-stained membranes (right). Scale bar is 2 μm. Fig. S6 contains larger fields of cells from the same experiment.
Septal peptidoglycan thinning is required for σG activity
We have shown that in sporulating cells lacking SpoIID and SpoIIP, GFP-SpoIIQ localizes to the septal membrane in a SpoIIIAH-dependent manner (Fig. 1). This suggests that SpoIIIAH and SpoIIQ are able to interact across the polar septum in the absence of PG thinning. We therefore wondered whether this interaction is sufficient to assemble an active secretion complex. We have previously shown that in cells lacking SpoIID, 11% of the sporulating cells maintain σG-dependent gene expression and therefore have active secretion complexes (Doan et al., 2009) (Fig. 7). However, sporulating cells lacking SpoIID have septal bulges that are thought to contain partially degraded PG raising the possibility that thinning of septal PG may be important for proper assembly and/or activity of the complex. To more directly test this idea, we analyzed σG activity in sporulating cells lacking both SpoIID and SpoIIP. In the absence of both cell wall hydrolases, the septal PG remains largely intact.
Figure 7. Septal peptidoglycan thinning by SpoIID and SpoIIP is required for σG activity.
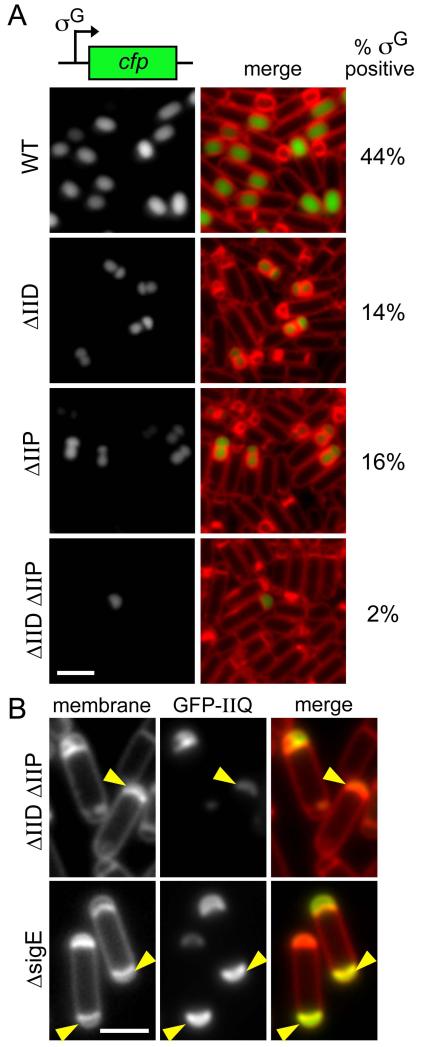
(A) Representative images of strains harboring the σG-responsive fluorescent reporter PsspE-cfp at hour 3 of sporulation. From top to bottom: wild-type (BTD2583), ΔspoIID (BTD2607), ΔspoIIP (BCR527), and a ΔspoIID, ΔspoIIP double mutant (BCR529). The percentages (n>500) of σG positive cells are indicated on the right. Images are CFP (left) and merge with TMA-DPH-stained membranes (right). Scale bar is 2 μm. Fig. S7 contains larger fields of cells from the same experiment. (B) Representative images of forespores at hour 3 of sporulation in ΔspoIID, ΔspoIIP (BKM1928) and ΔsigE (BKM1930). Examples of septal membranes that appear to have collapsed are indicated (yellow carets). Images are membranes stained with TMA-DPH (left), GFP-SpoIIQ (middle), and merged (right).
As described above, σG activity was assessed in single cells by fluorescence microscopy using a σG-responsive promoter (PsspE) fused to cfp (Doan et al., 2009). At hour 3 of sporulation, 44% of wild-type cells contained strong σG-dependent CFP fluorescence (Fig. 7A and S7). Similar to and in extension of what was reported previously, at the same timepoint, 14% and 16% of the sporulating cells lacking SpoIID or SpoIIP, respectively, contained strong σG-dependent forespore fluorescence. Furthermore, those forespores with σG activity also contained septal bulges (Fig. 7A and S7). Consistent with the idea that septal PG degradation is necessary to assemble active secretion complexes, in the ΔspoIID, ΔspoIIP double mutant, septal bulging was rarely observed and only a small fraction of the sporulating cells (2%) contained σG activity. The few cells that had σG-dependent fluorescence appeared to exhibit some bulging and/or to have deformed septal membranes (data not shown). Moreover, the fluorescence signal observed in these cells was weaker than in wild-type and in the single mutants, indicating a lower level of σG activity.
If, as our data suggest, the secretion complex is inactive in the absence of PG thinning, then the forespores should collapse in the ΔspoIID, ΔspoIIP double mutant. Although it is unclear what a collapsed forespore would look like in the absence of engulfment, we note that at hour 3 of sporulation (when forespores have begun to collapse in cells lacking SpoIIQ or SpoIIIA proteins), the septal membranes in the ΔspoIID, ΔspoIIP mutant bulge inward toward the forespore cell pole (Fig. 7B). Cells lacking σE (and do not produce SpoIID, SpoIIP and the SpoIIIA proteins) have a similar forespore phenotype (Fig. 7B). Interestingly, some of the early electron micrographs of mutants that lack σE activity reveal a phenotype similar to the one described here (Illing & Errington, 1991). We hypothesize that this is indeed “forespore collapse” and is a manifestation of the loss of metabolic potential. Altogether, these data suggest that SpoIID- and SpoIIP-mediated septal thinning is critical to assemble an active secretion complex.
Discussion
We have shown that the localization of SpoIIQ to the septal membrane on the forespore side depends on the mother cell protein SpoIIIAH and the thinning of the septal PG during engulfment. Our data further suggest that PG degradation allows an additional mother-cell protein to interact with and participate in anchoring SpoIIQ in the apposing membrane. Finally, our results support a model in which PG degradation is critical to assemble an active transenvelope secretion complex and that this second SpoIIQ-interacting protein functions with SpoIIIAH in its assembly (Figure 8). Type III secretion systems found in Gram-negative bacteria typically possess 20-25 proteins, of which 9-10 are highly conserved core components (Abby & Rocha, 2012, Buttner, 2012, Diepold et al., 2010, Ghosh, 2004). Type II and Type IV secretion systems are generally composed of 12-16 proteins (Filloux, 2004, Johnson et al., 2006, Korotkov et al., 2012, McLaughlin et al., 2012, Douzi et al., 2012, Christie et al., 2005, Fronzes et al., 2009, Chandran, 2013), of which a subset are highly conserved and likely to serve as the core machinery. Our findings raise the possibility that there are many unidentified proteins that similarly participate in the assembly and activity of this Gram-positive specialized secretion machine. Finally, our data highlight similar challenges and solutions that these macromolecular complexes must attend to in traversing the cell wall.
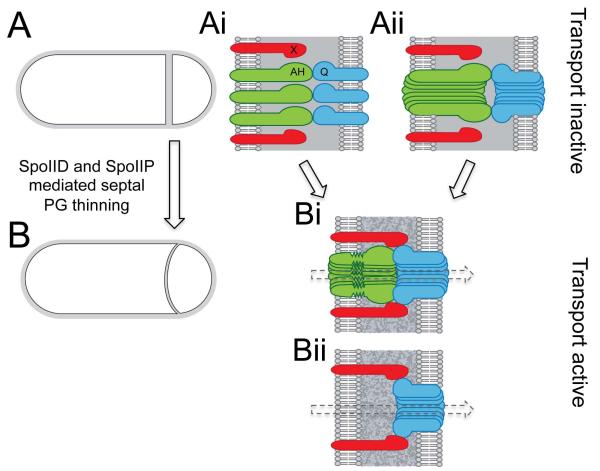
Figure 8. Septal peptidoglycan thinning facilitates the assembly and activity of the transenvelope secretion complex.
(A) Schematic representation of a sporulating cell with a flat asymmetric septum that has yet to undergo thinning of the septal PG (grey). (Ai and Aii) SpoIIQ (blue) and SpoIIIAH (green) can interact across the septal PG. By contrast, septal PG or the distance between the two membranes (~40 nm) prevents the interaction between SpoIIQ and its other mother-cell partner protein (X, in red). In the absence of septal thinning, the transenvelope secretion complex is not properly assembled (Ai) and/or inactive (Aii) resulting in forespore collapse and loss of σG activity. (B) Schematic representation of a sporulating cell that has undergone septal PG thinning (speckled grey). (Bi and Bii) Hydrolysis of the septal PG layers decreases the distance between the mother-cell and forespore membranes (~20 nm) and facilitates the interaction between SpoIIQ and its other mother cell partner. Septal PG thinning also results in active transport and σG activity. (Bi) SpoIIIAH and/or SpoIIQ adopt an altered conformation as a result of the reduced distance between the two membranes. (Bii) In the absence of SpoIIIAH, the secretion apparatus retains some activity due to the interaction between SpoIIQ and its other mother cell partner. For simplicity, the other SpoIIIA proteins in the secretion apparatus are not shown.
Although our data cannot formally rule out the possibility that the second anchor for SpoIIQ is modified PG, we favor a model in which this anchor is a mother-cell-produced protein that interacts directly with SpoIIQ. This is based on our inability to detect an interaction between the extracellular domain of SpoIIQ and PG in a collection of different assays using purified cell wall from vegetative and sporulating B. subtilis cells, in addition to E. coli sacculi. Furthermore, our data showing that induction of SpoIID, SpoIIP and SpoIIM in cells lacking σE argue that if there is a modification to septal PG that SpoIIQ recognizes, it is not generated by SpoIID and SpoIIP but yet another cell wall modifying enzyme activated by σE. Alternatively, cell wall synthesis under σE control could generate new PG that SpoIIQ binds. We do not favor this model because GFP-SpoIIQ remained properly localized in strains lacking SpoIIIAH and the cell wall synthesis machinery (SpoVD and SpoVE) (Daniel et al., 1994, Henriques et al., 1992, Fay et al., 2010, Meyer et al., 2010) under σE control (data not shown). Accordingly, we favor a simpler model that the second anchor for SpoIIQ is a protein synthesized in the mother, and that this protein unlike SpoIIIAH, requires the removal of septal PG to stably interact with SpoIIQ (Fig. 8). Importantly, this unidentified protein is not encoded in the spoIIIA operon, as GFP-SpoIIQ localization in a strain deleted for the entire operon was indistinguishable from the spoIIIAH null (Fig. 1).
As described in the Introduction, experiments from several groups suggest that SpoIIQ and SpoIIIAH form the basement layer (or scaffold) of the transenvelope secretion apparatus (Camp & Losick, 2008, Camp & Losick, 2009, Meisner et al., 2008, Doan et al., 2009). One observation that appeared in conflict with this model was the different requirement for these proteins in spore formation and in forespore gene expression under σG control. Cells lacking SpoIIQ or any of the SpoIIIA proteins, except SpoIIIAH, are severely impaired in sporulation (reduced 1000-fold) (Doan et al., 2009, Camp & Losick, 2008). By contrast, cells lacking SpoIIIAH have a mild defect in spore-formation (reduced 10-20-fold). Similarly, 10-20-fold more forespores exhibit σG-dependent gene expression in the SpoIIIAH mutant compared to mutants in SpoIIQ or the other SpoIIIA proteins (Doan et al., 2009) (Fig. 6B). The work presented here provides an explanation for this apparent paradox. Our data suggest that SpoIIIAH and a second mother-cell anchoring protein function together in building the transenvelope machine. Specifically, we imagine that in the absence of SpoIIIAH this second protein interacts with SpoIIQ across the double membrane septum and helps assemble the secretion complex (Fig. 8). The data presented here and previous localization studies in strains lacking SpoIIIAH (Doan et al., 2009) suggest that in the absence of SpoIIIAH fewer complexes form. However, those that do, presumably mediated by the second SpoIIQ anchor, appear to be sufficient to maintain forespore integrity and support σG activity and spore formation. A central challenge for the future is identifying this second mother-cell anchoring protein.
A role for the LytM groove in protein-protein interaction
The LytM domain was originally characterized on the lystostaphin D,D-endopeptidase from Staphyloccocus aureus and is a Zn2+ metallopeptidase (Odintsov et al., 2004). A large number of proteins contain this domain or a degenerate version that lacks the conserved residues involved in Zn2+ coordination. In the case of SpoIIQ, some of these residues are present but are not required for any of the known activities of this protein (Camp & Losick, 2009). The work presented here suggests that these domains could also serve as a protein-protein interaction surface. We suspect that many of the degenerate LytM domains could function in this capacity.
A role for SpoIID and SpoIIP in facilitating intermembrane protein-protein interaction and assembly of an active transenvelope complex
In addition to their role in promoting engulfment, this work and previous studies by the groups of Pogliano and Moran (Blaylock et al., 2004) reveal that SpoIID and SpoIIP play an important role in facilitating protein-protein interaction across the septum and in the assembly of an active secretion apparatus. Consistent with the idea that the SpoIIQ-SpoIIIAH interaction is weaker in the presence of septal PG, the septal localization of both proteins is reduced in cells lacking SpoIID and SpoIIP (Blaylock et al., 2004) (Fig. 1). Moreover, our data suggest that the second SpoIIQ anchoring protein is even more sensitive to the presence of the cell wall at the septum. One possibility is that this protein simply interacts more weakly with SpoIIQ. Alternatively, the extracytoplasmic domain of this protein could be shorter than that of SpoIIIAH and simply cannot “reach” across the space between the double-membrane prior to PG degradation (Fig. 8). Recent cryoelectron tomography of sporulating B. subtilis cells indicates that, prior to degradation of the septal PG, the distance between the two membranes is approximately 40 nm and after septal PG thinning, the intermembrane space is reduced to 20 nm (Tocheva et al., 2013). Thus, the proximity of the apposing membranes after PG degradation might be sufficient to allow the second mother cell protein to interact with SpoIIQ (Fig. 8). A final possibility is that the extracellular domain of this second protein might not fit through the pores in the cell wall that are thought to have a radius of ~2 nm (Demchick & Koch, 1996) and thus the PG would serve as a true barrier to their interaction. These scenarios are not mutually exclusive and could all contribute to a requirement for cell wall removal in anchoring SpoIIQ.
Finally, we envision two scenarios for how PG degradation might influence the activity of the secretion complex (Fig. 8). In the first, SpoIIQ and SpoIIIAH interact prior to PG degradation but are unable to assemble into a secretion channel (Fig. 8Ai). In the second, these proteins form a channel in the presence of the septal PG but other proteins cannot assemble into the complex or the machinery remains inactive until the PG is degraded (Fig. 8Aii and Bi). Based on the co-crystal structure, the extracytoplasmic domains of SpoIIIAH and SpoIIQ span approximately 5 nm. Yet, the distance between the two membranes, prior to PG degradation, is approximately 40 nm (Tocheva et al., 2013). Accordingly, for these proteins to interact, the regions of SpoIIIAH and SpoIIQ that connect the transmembrane segments to the folded domains, and that were disordered in the crystal structure (Levdikov et al., 2012), would have to adopt an extended conformation. Furthermore, upon PG thinning, these regions would need to accommodate the reduced space between the two membranes (Fig. 8Bi). We suspect that these changes in conformation are important in assembling an active complex and may also be critical to form the SpoIIIAH-SpoIIQ channel. Finally, as mentioned above, even after septal PG degradation the membranes remain ~20 nm apart (Tocheva et al., 2013). This suggests that additional proteins are required to make up the channel that connects the mother cell and forespore. SpoIIIAG and SpoIIIAF share weak similarity to SpoIIIAH and the YscJ/FliF family (Meisner et al., 2008, Doan et al., 2009) and are therefore good candidates to assemble into connected ring-like structures to span the double membrane as has been observed in Type III secretion systems (Marlovits et al., 2004, Schraidt et al., 2010).
A variety of specialized secretion systems that are organized into operons typically encode peptidoglycan-degrading enzymes (Scheurwater & Burrows, 2011). It has been suggested that those that lack these enzymes utilize PG hydrolases associated with general PG metabolism. None of the proteins encoded in the spoIIIA operon resembles a peptidoglycan-degrading enzyme. Accordingly, we hypothesize that this sporulation-specific secretion system has evolved to utilize SpoIID and SpoIIP as a means to accommodate the assembly of its components within the PG between the mother cell and forespore.
Materials and Methods
General methods
All B. subtilis strains were derived from the prototrophic strain PY79 (Youngman et al., 1983). Sporulation was induced by resuspension at 37°C according to the method of Sterlini-Mandelstam (Harwood & Cutting, 1990) or by exhaustion in supplemented DS medium (Schaeffer et al., 1965). Sporulation efficiency was determined in 24-30 hour cultures as the total number of heat-resistant (80°C for 20 min) colony forming units (CFUs) compared with wild-type heat-resistant CFUs. Induction of spoIID, spoIIP, and spoIIM during the early stages of sporulation was with 1 mM IPTG. Deletion mutants were generated by isothermal assembly (Gibson, 2011) and direct transformation of assembled DNA products into B. subtilis. Tables of strains, plasmids and oligonucleotide primers and descriptions of plasmid construction and isothermal assembly deletion mutants can be found online as supplementary material.
Immunoblot analysis
Whole-cell lysates from sporulating cells (induced by resuspension) were prepared as described previously (Doan et al., 2009). Samples were heated for 10 min at 50°C prior to loading. Equivalent loading was based on OD600 at the time of harvest. Proteins were separated by SDS-PAGE on 12.5% polyacrylamide gels, electroblotted onto Immobilon-P membranes (Millipore) and blocked in 5% nonfat milk in phosphate-buffered saline (PBS)-0.5% Tween-20. The blocked membranes were probed with anti-SpoIID (1:10,000) (Doan & Rudner, 2007), anti-SpoIIP (1:5,000) (Morlot et al., 2010), diluted into 3% BSA in 1X PBS-0.05% Tween-20. Primary antibodies were detected using horseradish peroxidase-conjugated goat, anti-rabbit IgG (BioRad) and the Western Lightning reagent kit as described by the manufacturer (PerkinElmer).
Fluorescence microscopy
Fluorescence microscopy was performed with an Olympus BX61 microscope as previously described (Doan et al., 2009). Cells were mounted on a 2% agarose pad containing resuspension medium using a gene frame (BioRad). Fluorescent signals were visualized with a phase contrast objective UplanF1 100x and captured with a monochrome CoolSnapHQ digital camera (Photometrics) using Metamorph software version 6.1 (Universal Imaging). The membrane dye TMA-DPH (Molecular Probes) was used at a final concentration of 0.01 mM and exposure times were typically 200 ms. Images were analyzed, adjusted and cropped using Metamorph software.
Quantification of σG positive cells
σG activity was assessed in single cells at 3 and 3.5 hours after the onset of sporulation by microscopy using fluorescent reporters (PsspB-cfp and PsspE-cfp) (Doan et al., 2009). For the experiments in Figure 6, a forespore was considered σG positive if it contained forespore fluorescence; it displayed normal forespore membrane morphology and was of normal size. The second and third criteria were included in the analysis to help assess σG activity in forespores with faint forespore fluorescence. Faint forespore fluorescence in a normal-sized forespore with unperturbed membranes was scored as σG positive. Faint forespore fluorescence in a small forespore with aberrant membrane morphology indicates forespore collapse (Doan et al., 2009) and was scored as σG negative. The percentage of σG positive cells was calculated based on the total number of cells that had completed engulfment. For the experiments in Figure 7, the cells examined were defective in engulfment. Accordingly, the percentage of σG positive cells was determined based on the total number of cells that had an asymmetric septum.
Supplementary Material
Acknowledgements
We thank members of the Rudner and Bernhardt labs for advice and encouragement, Leslie Wardwell for BLW002 and pLW003, Tsuyoshi Uehara for advice with protein and peptidoglycan purification and for purified EnvC, Catherine Paradis-Bleau for stimulating discussions, Tom Bernhardt for plasmids, and Rich Losick and Kit Pogliano for strains. Support for this work comes from the National Institute of Health Grant GM086466 (D.Z.R.).
REFERENCES
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abby SS, Rocha EP. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung S, Shum J, Abanes-De Mello A, Broder DH, Fredlund-Gutierrez J, Chiba S, Pogliano K. Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol Microbiol. 2007;65:1534–1546. doi: 10.1111/j.1365-2958.2007.05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP, Jr., Pogliano K. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol. 2008;69:402–417. doi: 10.1111/j.1365-2958.2008.06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009;23:1014, 1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V. Type IV secretion machinery: molecular architecture and function. Biochem Soc Trans. 2013;41:17–28. doi: 10.1042/BST20120332. [DOI] [PubMed] [Google Scholar]
- Chastanet A, Losick R. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol Microbiol. 2007;64:139–152. doi: 10.1111/j.1365-2958.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Drake S, Buchanan CE, Scholle R, Errington J. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J Mol Biol. 1994;235:209–220. doi: 10.1016/s0022-2836(05)80027-0. [DOI] [PubMed] [Google Scholar]
- Demchick P, Koch AL. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J Bacteriol. 1996;178:768–773. doi: 10.1128/jb.178.3.768-773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A, Amstutz M, Abel S, Sorg I, Jenal U, Cornelis GR. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 2010;29:1928–1940. doi: 10.1038/emboj.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Jr., Rudner DZ. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009;5:e1000566. doi: 10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Rudner DZ. Perturbations to engulfment trigger a degradative response that prevents cell-cell signalling during sporulation in Bacillus subtilis. Mol Microbiol. 2007;64:500–511. doi: 10.1111/j.1365-2958.2007.05677.x. [DOI] [PubMed] [Google Scholar]
- Douzi B, Filloux A, Voulhoux R. On the path to uncover the bacterial type II secretion system. Philos Trans R Soc Lond B Biol Sci. 2012;367:1059–1072. doi: 10.1098/rstb.2011.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger P, Fawcett P, Losick R. A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol Microbiol. 2001;42:1147–1162. doi: 10.1046/j.1365-2958.2001.02660.x. [DOI] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Fay A, Meyer P, Dworkin J. Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J Mol Biol. 2010;399:547–561. doi: 10.1016/j.jmb.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A. The underlying mechanisms of type II protein secretion. Biochim Biophys Acta. 2004;1694:163–179. doi: 10.1016/j.bbamcr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Fredlund J, Broder D, Fleming T, Claussin C, Pogliano K. The SpoIIQ landmark protein has different requirements for septal localization and immobilization. Mol Microbiol. 2013;XXX:XXXX–XX. doi: 10.1111/mmi.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev. 2004;68:771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez J, Smith R, Pogliano K. SpoIID-mediated peptidoglycan degradation is required throughout engulfment during Bacillus subtilis sporulation. J Bacteriol. 2010;192:3174–3186. doi: 10.1128/JB.00127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. Wiley; New York: 1990. [Google Scholar]
- Henriques AO, de Lencastre H, Piggot PJ. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie. 1992;74:735–748. doi: 10.1016/0300-9084(92)90146-6. [DOI] [PubMed] [Google Scholar]
- Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev. 2004;68:234–262. doi: 10.1128/MMBR.68.2.234-262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N, Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: roles of sigma E and sigma F in prespore engulfment. J Bacteriol. 1991;173:3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: from structure to function. FEMS Microbiol Lett. 2006;255:175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- Korotkov KV, Sandkvist M, Hol WG. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdikov VM, Blagova EV, McFeat A, Fogg MJ, Wilson KS, Wilkinson AJ. Structure of components of an intercellular channel complex in sporulating Bacillus subtilis. Proc Natl Acad Sci U S A. 2012;109:5441–5445. doi: 10.1073/pnas.1120087109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin LS, Haft RJ, Forest KT. Structural insights into the Type II secretion nanomachine. Curr Opin Struct Biol. 2012;22:208–216. doi: 10.1016/j.sbi.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Maehigashi T, Andre I, Dunham CM, Moran CP., Jr. Structure of the basal components of a bacterial transporter. Proc Natl Acad Sci U S A. 2012;109:5446–5451. doi: 10.1073/pnas.1120113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Moran CP., Jr. A LytM domain dictates the localization of proteins to the mother cell-forespore interface during bacterial endospore formation. J Bacteriol. 2011;193:591–598. doi: 10.1128/JB.01270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Wang X, Serrano M, Henriques AO, Moran CP., Jr. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci U S A. 2008;105:15100–15105. doi: 10.1073/pnas.0806301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, Chudakov DM. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- Meyer P, Gutierrez J, Pogliano K, Dworkin J. Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol Microbiol. 2010;76:956–970. doi: 10.1111/j.1365-2958.2010.07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev. 2010;24:411–422. doi: 10.1101/gad.1878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odintsov SG, Sabala I, Marcyjaniak M, Bochtler M. Latent LytM at 1.3A resolution. J Mol Biol. 2004;335:775–785. doi: 10.1016/j.jmb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Perez AR, Abanes-De Mello A, Pogliano K. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in bacillus subtilis. J Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A, Pogliano K. Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis. EMBO J. 2004;23:1636–1646. doi: 10.1038/sj.emboj.7600171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabala I, Jonsson IM, Tarkowski A, Bochtler M. Anti-staphylococcal activities of lysostaphin and LytM catalytic domain. BMC Microbiol. 2012;12:97. doi: 10.1186/1471-2180-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurwater EM, Burrows LL. Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol Lett. 2011;318:1–9. doi: 10.1111/j.1574-6968.2011.02228.x. [DOI] [PubMed] [Google Scholar]
- Schraidt O, Lefebre MD, Brunner MJ, Schmied WH, Schmidt A, Radics J, Mechtler K, Galan JE, Marlovits TC. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 2010;6:e1000824. doi: 10.1371/journal.ppat.1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shida T, Hattori H, Ise F, Sekiguchi J. Mutational analysis of catalytic sites of the cell wall lytic N-acetylmuramoyl-L-alanine amidases CwlC and CwlV. J Biol Chem. 2001;276:28140–28146. doi: 10.1074/jbc.M103903200. [DOI] [PubMed] [Google Scholar]
- Tocheva EI, Lopez-Garrido J, Hughes HV, Fredlund J, Kuru E, Vannieuwenhze MS, Brun YV, Pogliano K, Jensen GJ. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol. 2013 doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman PJ, Perkins JB, Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983;80:2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.