Abstract
BRAFV600E mutation in microsatellite unstable (MSI) CRCs virtually excludes Lynch Syndrome (LS). In microsatellite stable (MSS) CRC it predicts poor prognosis. We propose a universal CRC LS screening algorithm using concurrent reflex immunohistochemistry (IHC) for BRAFV600E and MMR proteins. We compared BRAFV600E IHC to multiplex polymerase chain reaction (PCR) and MALDI-TOF spectrometry in 216 consecutive CRCs from 2011. Discordant cases were resolved with rt-PCR. BRAFV600E IHC was performed on 51 CRCs from the Australasian Colorectal Cancer Family Registry (ACCFR) which were fully characterised for BRAF mutation by allele-specific PCR, MMR status (MMR IHC and MSI), MLH1 promoter methylation and germline MLH1 mutation. We then assessed MMR and BRAFV600E IHC on 1403 consecutive CRCs.
By MALDI-TOF 15 cases did not yield a BRAF result, while 38/201(19%) were positive. By IHC 45/216(20%) were positive. Of the 7 discordant cases, rt-PCR confirmed the IHC result in 6. In the 51 CRCs from the ACCFR, IHC was concordant with allele-specific PCR in 50 cases. BRAFV600E and MSI IHC on 1403 CRCs demonstrated the following phenotypes: BRAF-ve/MSS (1029 cases,73%), BRAF+ve/MSS (98,7%), BRAF+ve/MSI (183,13%), and BRAF-ve/MSI (93,7%). All 11/1403 cancers associated with proven LS were BRAF-ve/MSI.
We conclude that BRAF IHC is highly concordant with two commonly used PCR-based BRAFV600E assays, performed well in identifying MLH1 mutation carriers from the ACCFR and identified all cases of proven LS out of 1403 CRCs. Reflex BRAFV600E and MMR IHC are simple cheap tests which facilitate universal LS screening and identify the poor prognosis BRAFV600E mutant MSS CRC phenotype.
Introduction
Lynch Syndrome (LS) accounts for 2-4% of colorectal carcinoma (CRC) and is characterized by tumors with microsatellite instability (MSI) and loss of mismatch repair (MMR) protein expression due to germline mutations in MLH1, PMS2, MSH2 and MSH6.[1-4] Identification of LS facilitates screening and risk reduction strategies.[5] Therefore most centers have a screening strategy for patients presenting with CRC. Screening usually begins with either immunohistochemistry (IHC) for the four MMR enzymes MLH1, PMS2, MSH2 and MSH6 or PCR for MSI which are essentially equally effective.[2, 3, 6].
It has been debated whether MMR/MSI testing should be performed on all CRC or only on those with the ‘red flags’ of synchronous or metachronous tumors, family history, distinctive histology or young age of onset.[6, 7] There is now a trend towards widespread screening strategies including those which recommend reflex IHC testing for all patients with CRC regardless of age or other risk factors.[7-9] Universal screening is more expensive but identifies significantly more cases of LS than targeted approaches - perhaps up to 28% of all cases of LS.[3]
Loss of expression of MLH1 and PMS2 is not limited to LS but also occurs due to somatic hyper-methylation of the MLH1 gene promoter causing transcriptional silencing of MLH1 in 10-5% of all CRC.[3] While PMS2 negative/MLH1 positive tumors or tumors with negative staining for MSH2 or MSH6 are highly correlated with LS at any age,[9] the proportion of CRC with somatic hyper-methylation of MLH1 (resulting in an MLH1 negative/PMS2 negative phenotype) increases with age as the absolute risk of LS decreases. It is the further investigation by molecular testing of this large number of CRC patients who have MLH1 silencing due to somatic hyper-methylation which adds significant extra downstream cost to universal LS screening by reflex MMR IHC.
Activating mutations in BRAF are found in 5-25% of CRC, with the vast majority being the BRAFV600E mutation.[10-14] BRAFV600E mutation occurs in two thirds of CRC with MLH1 silencing due to somatic hyper-methylation but virtually never in CRC with MSI due to LS.[4] Therefore BRAFV600E mutation is used as a proxy marker for hyper-methylation in MLH1 IHC negative tumors. Further testing for LS is commonly offered on MLH1 negative cases only if they are BRAF wild type.[9]
BRAFV600E mutation also occurs in MSS tumors. This group has emerged as a distinct molecular and clinical phenotype with a poor prognosis.[12, 15-22] BRAFV600E mutation is mutually exclusive with KRAS mutation but its presence may predict a worse or absent response to EGFR inhibition.[23] Therefore the American National Comprehensive Cancer Network guidelines now recommend consideration of BRAF testing to guide therapy in the setting of KRAS wild type metastatic CRC.[24] Assessment of BRAF status in all CRC patients may provide useful therapeutic and prognostic information but its routine assessment is justified only if it can be delivered cheaply and efficiently.
Recently we developed a novel mouse monoclonal mutation specific anti-BRAFV600E antibody (clone VE1) which can be used for IHC on routinely processed formalin fixed paraffin embedded (FFPE) tissue.[25] This antibody reacts with the protein produced by the BRAFV600E mutation but not with wild type BRAF or other BRAF mutants and has now been shown to be robust and reliable by several groups in several different tumors including papillary thyroid cancer, melanoma, cerebral neoplasia, ovarian tumors and hairy cell leukemia.[25-32] Importantly BRAF IHC can potentially be performed in any diagnostic pathology laboratory which currently offers MMR IHC. Given the workflow patterns in diagnostic pathology laboratories, BRAF IHC can be performed concurrently with MMR IHC at little extra cost. It is simply a matter of performing IHC for five markers rather than four.
In this study, we validate an IHC test for the BRAFV600E mutation in CRC, compare it to current PCR-based approaches for the detection of MLH1 mutation carriers and propose a new approach to screen for LS using reflex MMR and BRAFV600E mutation specific IHC.
Methods
We searched the pathology database of Royal North Shore Hospital, Sydney, for all cases of CRC treated by surgical resection during the period 2006 to 2011. Exclusion criteria included extra-colonic and appendiceal location, tumors treated endoluminally and histological type other than adenocarcinoma as defined by the WHO 2010 system.[33] Tumors were independently reviewed by two pathologists (CT and AG) to confirm the diagnosis and to reclassify the pathological stage according to the 7th edition 2009 AJCC/TNM system.[34] For resections involving synchronous tumors, the tumor with the highest pathological stage was selected and annotated. Tissue microarrays (TMA) comprising duplicate 1mm diameter cores were created from formalin fixed paraffin embedded (FFPE) tissue blocks.
IHC for BRAFV600E was performed on FFPE whole sections from all available CRC cases from the year 2011 using a commercially available mouse monoclonal anti-BRAFV600E antibody (clone VE1; provided by AvD and DC; available at SpringBioscience Pleasonton CA USA).[25] Cases with any positive IHC staining (defined as diffuse strong positive staining of >75% of malignant cells) were scored as positive. IHC scoring was performed independently by three pathologists (AG, CT and AC) blinded to all clinical, molecular and pathological data. Discordant scores were resolved by consensus review.
Molecular testing for BRAFV600E mutation was performed on macro-dissected tumor tissue from the same block used for IHC using a multiplex PCR and MALDI-TOF mass spectrometry detection assay (Sequenom MassArray).[35] This platform has been specifically validated for BRAFV600E, V600R, V600K and V600M and the laboratory holds full National Association of Testing Authorities Australia accreditation for this assay. The paraffin blocks of discordant cases were macro-dissected a second time and re-analyzed using a real-time PCR (rt-PCR) based assay (Roche COBAS 4800 BRAF V600 Mutation Test). All molecular testing was performed blinded to clinical, pathological and IHC data.
IHC for BRAFV600E and the four MMR proteins (MLH1, PMS2, MSH2 and MSH6) was performed on the TMA slides for the entire cohort of 2006 to 2011. The accuracy of BRAFV600E IHC on the TMA was validated by comparison with whole section IHC and molecular testing performed on the cases from the year 2011. TMA IHC scoring was performed independently by two pathologists (AG and AC). Discordant cases were resolved by a third pathologist (CT). All assessors were blinded to clinical, molecular and pathological data.
To determine whether all known LS cases demonstrated a BRAF-ve/MSI phenotype, the files of the hospital familial cancer clinic were searched for all patients with genetically proven LS who underwent surgery for CRC during this period.
To further investigate the role for BRAF IHC in triaging formal genetic testing in patients with suspected LS based on negative staining for MLH1 and PMS2 in an external cohort processed in different laboratories, IHC was performed on archived FFPE tissue blocks of cases from the Jeremy Jass Memorial Tissue Bank of the Australasian-Colorectal-Cancer-Family-Registry (ACCFR). This cohort includes CRCs processed in multiple different laboratories with different fixation and processing approaches collected over a period of years. The ACCFR has been previously described, comprising a richly annotated cohort which includes MMR IHC information correlated with molecular data such as MSI status, MMR gene mutation status, MLH1 and CpG island methylator phenotype (CIMP) methylation analysis and BRAFV600E status determined by allele-specific PCR (AS-PCR).[36-38]. For this arm of the study CRC samples demonstrating loss of expression for MSH2 and/or MSH6 or solitary loss of PMS2 protein expression by IHC were excluded because of their very strong association with LS. IHC was interpreted blinded to all other data independently by AG and CT.
Statistical analyses were performed using IBM SPSS Statistics v20 (detailed methods presented in supplementary methods). The following measures of test performance were determined - positive percent agreement (PPA), negative percent agreement (NPA) and overall percent agreement (OPA).
This study was approved by the ethics committees of the participating institutions. Patients who underwent genetic testing provided informed consent.
Results
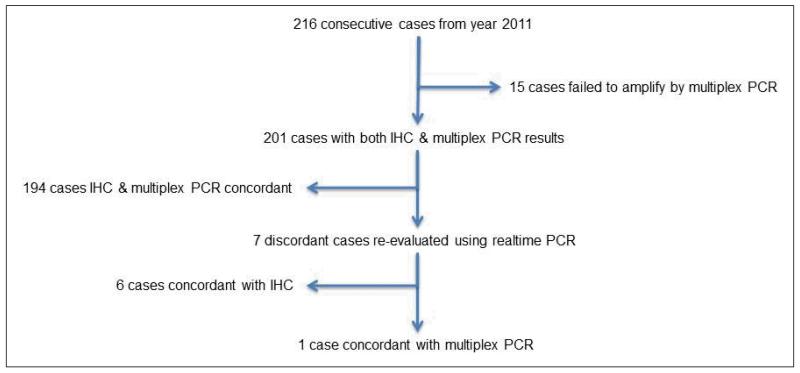
A search of all resected CRC from 2006 to 2011 yielded 1456 cases. After excluding cases which did not have at least one tumor core on TMA, 1403 cases were available for BRAF and MMR IHC. All CRCs resected in the calendar year 2011 (n=216) were used to validate BRAF IHC against the multiplex PCR and MassArray Spectrometry-based assay. The comparison of molecular analysis and BRAF IHC on the cases from 2011 is summarized in figure 1 and supplementary table 1. Briefly, 15 cases failed to amplify or provide a result by MassArray leaving 201 cases with both IHC and molecular results. No mutations other than V600E were detected by PCR. There was complete agreement between IHC and MassArray in all but 7 cases (6 negative by PCR but positive by IHC and 1 positive by PCR but negative by IHC). When these 7 discordant cases were re-evaluated using rt-PCR, all cases were positive. That is, rt-PCR favored the IHC result in 6 of 7 discordant cases. IHC was repeated on the apparently false negative case a number of times and yielded the same result. It was noted that this case was predominantly composed of signet ring cells with little non-mucinous cytoplasm.
Figure 1.
Comparison of BRAFV600E IHC and PCR in 216 consecutive cases from the year 2011.
BRAF IHC on whole sections was readily interpretable by experienced observers. Although we chose the arbitrary cut-off of requiring 75% of malignant cells to show staining to be considered positive, the great majority of positive cases actually demonstrated diffuse strong homogeneous cytoplasmic staining in essentially all malignant cells while the great majority of negative cases showed completely absent staining in all malignant cells (figure 2). Patchy non-specific staining was not uncommonly seen in some smooth muscle cells, mucin and non-neoplastic colonic mucosa (sometimes with a peculiar nuclear pattern of staining). Occasional positive cases demonstrated only weak but still quite diffuse cytoplasmic staining. In these positive cases the pattern of staining in the neoplastic cells was still quite distinct and different to any non-specific staining seen in the non-neoplastic cells.
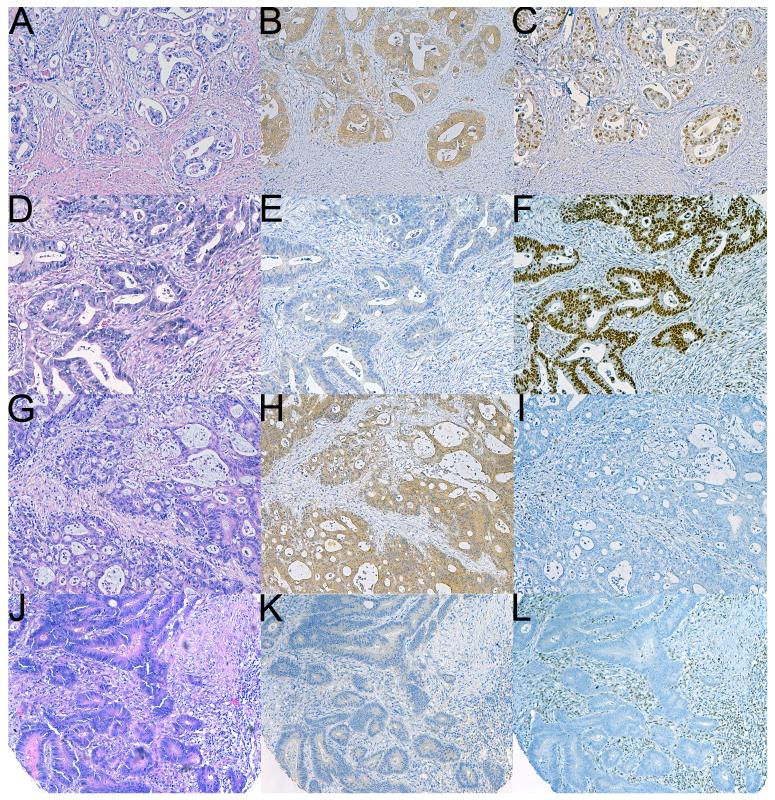
Figure 2.
Serial sections of four different CRCs stained with hematoxylin and eosin (A,D,G,J), and IHC for BRAFV600E (B,E,H,K) and MLH1 (C,F,I,L). The tumor in A-B-C is MSS (positive staining was also seen for PMS2, MSH2 and MSH6) and BRAFV600E positive. This appears to represent a poor prognostic group. The tumour in D-E-F is BRAFV600E negative and MSS. The tumor illustrated in G-H-I shows negative staining for MLH1 and demonstrates MSI. It also demonstrates positive staining for BRAFV600E therefore LS is essentially excluded. The tumor illustrated in J-K-L demonstrates MSI and negative staining for BRAFV600E therefore in this instance formal genetic testing for LS is justified. As illustrated in panels B and H, positive staining for BRAFV600E is characterized by widespread cytoplasmic staining limited to tumor cells (Original magnifications 100X).
When cases from 2011 stained on the TMA were compared to whole sections there were 5 discordant cases all of which were false negatives of the TMA attributed on review to heterogeneous staining. Compared to whole section IHC, BRAF IHC on TMA demonstrated lower positive predictive agreement, although negative predictive agreement and overall percent agreement were comparable (supplementary table 2).
The scoring of BRAF IHC on whole sections for the cases from 2011 was completely concordant between two observers (kappa score 1). The third observer only disagreed on one case (kappa score 0.985). BRAF IHC on the TMA of 1403 cases showed excellent concordance between two observers (kappa score 0.964). Scores for BRAF IHC on the ACCFR cohort were fully concordant between two observers (kappa score 1).
The clinical and pathological details of the full cohort of 1403 CRC cases assessed on TMA is summarized in supplementary table 3, supplementary figure 1 and table 1. Briefly, the prevalence of both BRAFV600E mutation and MSI was 20% with the following phenotypes recorded: BRAF-ve/MSS (1029 cases, 73%), BRAF+ve/MSS (98 cases, 7%), BRAF+ve/MSI (183 cases,13%), and BRAF-ve/MSI (93 cases, 7%). In MSS tumours, BRAFV600E mutation was significantly associated with higher histologic grade, higher overall stage and a predilection for the right colon (supplementary table 4). These differences were absent when BRAF status was compared in MSI tumors (supplementary table 5).
Table 1.
The clinical and pathological characteristics of the four phenotypes of colorectal carcinoma, as defined by IHC. The figures are calculated as percentages within each phenotype. Cumulative percentage figures may exceed 100% due to rounding up to one significant figure.
| BRAF+ve/MSS | BRAF+ve/MSI | BRAF-ve/MSI | BRAF-ve/MSS | |
|---|---|---|---|---|
| Total, No (%) | 98 (7%) | 183 (13%) | 93 (7%) | 1029 (73%) |
| Gender, No. (%) | ||||
| Female | 57 (58%) | 136 (74%) | 53 (57%) | 468 (46%) |
| Male | 41 (42%) | 47 (26%) | 40 (43%) | 561 (55%) |
| Anatomic site, No. (%) | ||||
| Cecum | 20 (20%) | 50 (28%) | 33 (36%) | 182 (18%) |
| Ascending colon | 31 (32%) | 64 (36%) | 25 (27%) | 110 (11%) |
| Transverse colon | 16 (16%) | 40 (23%) | 21 (23%) | 71 (7%) |
| Descending colon | 9 (9%) | 5 (3%) | 4 (4%) | 64 (6%) |
| Sigmoid colon | 10 (10%) | 14 (8%) | 6 (6%) | 276 (27%) |
| Rectum | 12 (12%) | 3 (2%) | 4 (4%) | 324 (32%) |
| Histologic grade, No. (%) | ||||
| Low grade | 57 (58%) | 101 (57%) | 63 (68%) | 887 (86%) |
| High grade | 41 (42%) | 77 (43%) | 30 (32%) | 142 (14%) |
| pT stage, No. (%) | ||||
| Tis | 0 (0%) | 0 (0%) | 0 (0%) | 1 (<1%) |
| T1 | 2 (2%) | 7 (4%) | 8 (9%) | 80 (8%) |
| T2 | 6 (6%) | 18 (10%) | 9 (10%) | 175 (17%) |
| T3 | 37 (38%) | 112 (63%) | 53 (57%) | 515 (50%) |
| T4a | 50 (51%) | 30 (17%) | 15 (16%) | 217 (21%) |
| T4b | 3 (3%) | 11 (6%) | 8 (9%) | 41 (4%) |
| pN stage, No. (%) | ||||
| N0 | 23 (24%) | 114 (64%) | 69 (74%) | 558 (54%) |
| N1a | 12 (12%) | 21 (12%) | 7 (8%) | 130 (13%) |
| N1b | 21 (22%) | 18 (10%) | 5 (5%) | 125 (12%) |
| N1c | 2 (2%) | 3 (2%) | 1 (1%) | 34 (3%) |
| N2a | 21 (22%) | 13 (7%) | 3 (3%) | 103 (10%) |
| N2b | 18 (19%) | 9 (5%) | 8 (9%) | 79 (8%) |
| pM stage, No. (%) | ||||
| Mx | 88 (91%) | 174 (98%) | 91 (98%) | 989 (96%) |
| M1a | 4 (4%) | 1 (<1%) | 0 (0%) | 20 (2%) |
| M1b | 5 (5%) | 3 (2%) | 2 (2%) | 20 (2%) |
| Overall stage, No. (%) | ||||
| 0 | 0 (0%) | 0 (0%) | 0 (0%) | 1 (<1%) |
| I | 4 (4%) | 23 (13%) | 14 (15%) | 199 (19%) |
| IIA | 11 (11%) | 78 (44%) | 43 (46%) | 282 (27%) |
| IIB | 8 (8%) | 7 (4%) | 9 (10%) | 56 (5%) |
| IIC | 2 (2%) | 6 (3%) | 3 (3%) | 10 (1%) |
| IIIA | 4 (4%) | 3 (2%) | 2 (2%) | 46 (5%) |
| IIIB | 31 (32%) | 43 (24%) | 9 (10%) | 267 (26%) |
| IIIC | 29 (30%) | 14 (8%) | 11 (12%) | 127 (12%) |
| IVA | 4 (4%) | 1 (<1%) | 0 (0%) | 20 (2%) |
| IVB | 5 (5%) | 3 (2%) | 2 (2%) | 21 (2%) |
The results of MMR IHC sub-categorized for BRAF staining are presented in supplementary table 6. Briefly, 69% of MLH1-ve/PMS2-ve cases showed positive staining for BRAFV600E, as did 57% of triple-negative cases (MLH1-ve, PMS2-ve, MSH6-ve). Other than for the triple-negative cases, no MSH6 negative case showed positive staining for BRAFV600E. Of note there were no cases which were MLH1 negative which were not also PMS2 negative and no cases which were MSH2 negative which were not also MSH6 negative. That is, IHC for only PMS2 and MSH6 would have detected all MMR negative CRCs.
Of the 1403 cases in the TMA cohort, 11 cancers from 10 patients were confirmed to be associated with LS by molecular testing performed during routine care. All of these cancers demonstrated a BRAF-ve/MSI phenotype (supplementary table 7).
There were 51 CRC cases from 49 individuals obtained from both population- and clinic-based recruitment from the ACCFR. Of the 51 CRCs, 39 (76%) demonstrated loss of staining for MLH1 and PMS2 by IHC, and 23 (45%) were positive for the BRAFV600E mutation by AS-PCR (Table 2). Of the 23 CRC demonstrating the BRAFV600E mutation, 1 was associated with MLH1 mutation (c.790+2dupT r.[678_790del, 678_884del] p.?), 11 with MLH1 methylation and the remaining 11 cases were MMR-proficient. The CRC from the individual with both MLH1 germline mutation and BRAFV600E mutation was methylated at the RUNX3, CACNA1G, SOCS1, NEUROG1 and IGF2 loci, thereby demonstrating high levels of CIMP (CIMP-H). BRAF IHC was concordant with BRAFV600E determined by AS-PCR in all but one case, including the LS case with BRAFV600E mutation. The discordant case was positive by BRAF IHC but negative by AS-PCR and occurred in a patient with LS (MLH1 mutation c.678-1G>C r.spl? p.?). It did not demonstrate hyper-methylation of the MLH1 gene promoter or a CIMP-H phenotype. Repeat testing of both the AS-PCR and BRAF IHC from freshly cut sections and extracted DNA from the same block did not change the findings of either test.
Table 2.
Molecular and IHC characteristics of the 51 CRCs from the Australian Colorectal Cancer Familial Registry (ACCFR).
| Case | Individual | Category | BRAF IHC | BRAF AS-PCR | Region tested | Sequencing | MLPA | MMR IHC | MSI Class | MLH1 methylation | CIMP Overall | Primary_Site |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | Lynch Syndrome | negative | wildtype | MLH1 c.350C>T p.Thr117Met | MLH1 | no | MLH1/PMS2 loss | MSI-H | descending colon | ||
| 2 | 2 | Lynch Syndrome | negative | wildtype | MLH1 c.350C>T p.Thr117Met | MLH1 | no | MLH1/PMS2 loss | MSI-H | |||
| 3 | 3 | Lynch Syndrome | positive | wildtype | MLH1 c.678-1G>C r.spl? p.? | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | no methylation | negative | caecum |
| 4 | 4 | Lynch Syndrome | negative | wildtype | MLH1c.1852_1854delAAG p.Lys618del | MLH1 | no | MLH1/PMS2 loss | MSI-H | |||
| 5 | 5 | Lynch Syndrome | negative | wildtype | MLH1 c.790+2dupT r.[678_790del, 678_884del] p.? | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | caecum | ||
| 6 | 5 | Lynch Syndrome | negative | wildtype | MLH1 c.790+2dupT r.[678_790del, 678_884del] p.? | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | |||
| 7 | 6 | Lynch Syndrome | negative | wildtype | MLH1 del x3 | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | transverse colon | ||
| 8 | 7 | Lynch Syndrome | negative | wildtype | MLH1 c.380+1G>A r.spl? p.? | MLH1 | no | MLH1/PMS2 loss | MSI-H | ascending colon | ||
| 9 | 8 | Lynch Syndrome | negative | wildtype | MLH1 c.1758delC p.Ala586fs | MLH1 | no | MLH1/PMS2 loss | MSI-H | transverse colon | ||
| 10 | 9 | Lynch Syndrome | positive | V600E positive | MLH1 c.790+2dupT r.[678_790del, 678_884del] p.? | MLH1 | no | MLH1/PMS2 loss | MSI-H | no methylation | positive | transverse colon |
| 11 | 10 | Lynch Syndrome | negative | wildtype | MLH1 c.116+5G>C r.[116_117ins227, spl?] p.? | MLH1 | no | MLH1/PMS2 loss | MSI-H | ascending colon | ||
| 12 | 11 | Lynch Syndrome | negative | wildtype | MLH1 c.350C>T p.Thr117Met | MLH1 | no | MLH1/PMS2 loss | MSI-H | transverse colon | ||
| 13 | 12 | Lynch Syndrome | negative | wildtype | MLH1 c.1559-2A>T r.spl? p.? | MLH1 | no | MLH1/PMS2 loss | MSI-H | sigmoid colon | ||
| 14 | 13 | Lynch Syndrome | negative | wildtype | MLH1 c.2224delC p.Gln742SerfsextX*25 | MLH1 | no | MLH1/PMS2 loss | caecum | |||
| 15 | 14 | Lynch Syndrome | negative | wildtype | MLH1 c.1464_1468delGGAAA p.Lys488AspfsX13 | MLH1 | MLH1 | MLH1/PMS2 loss | no methylation | negative | transverse colon | |
| 16 | 15 | Lynch Syndrome | negative | wildtype | MLH1 c.672delT p.Ser225ValfsX4 | MLH1 | MLH1 | MLH1/PMS2 loss | no methylation | negative | caecum | |
| 17 | 16 | Lynch Syndrome | negative | wildtype | MLH1 c.554T>G p.Val185Gly | MLH1 | MLH1 | MLH1/PMS2 loss | no methylation | negative | descending colon | |
| 18 | 17 | Lynch Syndrome | negative | wildtype | MLH1 c.76C>T p.Gln26X | MLH1 | no | MLH1/PMS2 loss | no methylation | negative | transverse colon | |
| 19 | 18 | Lynch Syndrome | negative | wildtype | MLH1 c.116+5G>C r.[116_117ins227, spl?] p.? | MLH1 | no | MLH1/PMS2 loss | MSI-L | caecum | ||
| 20 | 19 | Lynch Syndrome | negative | wildtype | MLH1 c.790+10A>G | MLH1 | MLH1/PMS2 loss | descending colon | ||||
| 21 | 20 | BRAF MSI-H | positive | V600E positive | No mutation found | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | positive | caecum |
| 22 | 21 | BRAF MSI-H | positive | V600E positive | No mutation found | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | positive | caecum |
| 23 | 22 | BRAF MSI-H | positive | V600E positive | no blood DNA for testing | no | no | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | positive | caecum |
| 24 | 23 | BRAF MSI-H | positive | V600E positive | No mutation found | MLH1 | no | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | positive | ascending colon |
| 25 | 24 | BRAF MSI-H | positive | V600E positive | no blood DNA for testing | no | no | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | positive | ascending colon |
| 26 | 25 | BRAF MSI-H | positive | V600E positive | no blood DNA for testing | no | no | MLH1/PMS2 loss | MLH1 methylation positive | positive | ascending colon | |
| 27 | 26 | BRAF MSI-H | positive | V600E positive | No mutation found | MLH1 | MLH1 | MLH1/PMS2 loss | MLH1 methylation positive | positive | caecum | |
| 28 | 27 | BRAF MSI-H | positive | V600E positive | No mutation found | no | no | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | positive | |
| 29 | 28 | BRAF MSI-H | positive | V600E positive | no blood DNA for testing | no | no | MLH1/PMS2 loss | MLH1 methylation positive | positive | caecum | |
| 30 | 28 | BRAF MSI-H | positive | V600E positive | no blood DNA for testing | no | no | MLH1/PMS2 loss | MLH1 methylation positive | positive | transverse colon | |
| 31 | 29 | BRAF MSI-H | positive | V600E positive | no blood DNA for testing | no | no | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | positive | sigmoid colon |
| 32 | 30 | BRAF MSS | positive | V600E positive | not tested yet | no | no | normal expression | MSI-L | no methylation | sigmoid colon | |
| 33 | 31 | BRAF MSS | positive | V600E positive | not tested yet | no | no | normal expression | MSS | rectum | ||
| 34 | 32 | BRAF MSS | positive | V600E positive | no blood DNA for testing | no | no | normal expression | MSS | sigmoid colon | ||
| 35 | 33 | BRAF MSS | positive | V600E positive | not tested yet | no | no | normal expression | MSS | caecum | ||
| 36 | 34 | BRAF MSS | positive | V600E positive | not tested yet | no | no | normal expression | MSS | caecum | ||
| 37 | 35 | BRAF MSS | positive | V600E positive | No mutation found | MLH1 | no | normal expression | MSI-L | descending colon | ||
| 38 | 36 | BRAF MSS | positive | V600E positive | No mutation found | MLH1 | no | normal expression | MSI-L | descending colon | ||
| 39 | 37 | BRAF MSS | positive | V600E positive | No mutation found | MLH1 | no | normal expression | MSS | no methylation | positive | sigmoid colon |
| 40 | 38 | BRAF MSS | positive | V600E positive | not tested yet | no | no | normal expression | MSS | transverse colon | ||
| 41 | 39 | BRAF MSS | positive | V600E positive | not tested yet | no | no | normal expression | MSS | rectum | ||
| 42 | 40 | BRAF MSS | positive | V600E positive | not tested yet | no | no | normal expression | MSS | no methylation | negative | rectum |
| 43 | 41 | MSI-H BRAFwt | negative | wildtype | No mutation found | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | no methylation | negative | ascending colon |
| 44 | 42 | MSI-H BRAFwt | negative | wildtype | No mutation found | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | positive | transverse colon |
| 45 | 43 | MSI-H BRAFwt | negative | wildtype | No mutation found | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | negative | caecum |
| 46 | 44 | MSI-H BRAFwt | negative | wildtype | no blood DNA for testing | no | no | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | positive | caecum |
| 47 | 45 | MSI-H BRAFwt | negative | wildtype | No mutation found | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | negative | transverse colon |
| 48 | 46 | MSI-H BRAFwt | negative | wildtype | No mutation found | MLH1 | MLH1 | MLH1/PMS2 loss | MSI-H | MLH1 methylation positive | negative | ascending colon |
| 49 | 47 | MSI-H BRAFwt | negative | wildtype | No mutation found | MLH1 | MLH1 | MLH1/PMS2 loss | MLH1 methylation positive | negative | caecum | |
| 50 | 48 | MSI-H BRAFwt | negative | wildtype | No mutation found | MLH1 | MLH1 | MLH1/PMS2 loss | MLH1 methylation positive | negative | rectum | |
| 51 | 49 | MSS BRAF wt | negative | wildtype | No mutation found | MLH1 | MLH1 | normal expression | MSS | rectum |
Discussion
Personalized medicine strategies and increased recognition of familial cancer syndromes have created an urgent need for rapid and accurate molecular characterization of cancers in the clinical diagnostic setting. Many techniques have been developed to identify BRAF mutations. These methods are all PCR based, relatively time consuming, require significant infrastructure, are technically demanding and subject to tissue heterogeneity, sampling error and suboptimal preservation of DNA in formalin fixed paraffin embedded (FFPE) tissue. For these reasons, BRAFV600E testing has been difficult to deploy clinically particularly in resource poor settings.
In this study we have demonstrated that simple IHC performed on routinely processed FFPE tissue in a standard surgical pathology laboratory compares favorably to two different but commonly used molecular platforms. There was high agreement between IHC and a multiplex PCR-based assay in the determination of BRAF status on FFPE tissue (PPA=97.4%, 95%CI=86.5-99.5%; NPA=96.3%, 95%CI=92.2-98.3%; OPA=96.5%, 95%CI=93.0-98.3%). These figures are comparable to published data comparing an allele-specific PCR based assay to sequencing (PPA=93.3%, 95%CI=66.0-99.7%)[30] and rt-PCR based assay (Roche COBAS 4800 BRAF Mutation test kit) to sequencing (PPA=96.4%, 95%CI=93.1-98.2%; NPA=80.0, 95%CI=74.1-84.8%; OPA=88.5, 95%CI=85.1-91.1%).[39]
In fact our results suggest that BRAF IHC may outperform PCR MassArray in the routine clinical setting. For example IHC provided a result in all cases (versus a failure rate of 7% for MassArray) and, using rt-PCR as the gold standard in discordant cases, IHC provided the ‘correct’ result in 200 of 201 cases (99.5%) whereas MassArray was ‘correct’ in only 195 of 201 cases (97%).
The BRAF antibody we used only reacts with the protein product of the V600E mutant and not with proteins associated with other mutations of BRAF.[25] This makes it ideal for use in CRC because BRAF mutations other than V600E are rare and not associated with somatic hyper-methylation.[10-14]
Royal North Shore Hospital in Sydney, Australia, performs centralized pathology testing for two quaternary referral hospitals with dedicated colorectal surgery units as well as four community hospitals. Therefore the cases processed at this center represent a true snapshot of CRC in the community rather than being biased towards the patient populations commonly seen in study cohorts. In conjunction with the increased sensitivity of BRAFV600E IHC, this older unselected population with its tendency to somatic hyper-methylation may account for our relatively high BRAF mutation rate (20%) and low rate of confirmed LS (0.8%).[10-14] It is noteworthy that even in this cohort in which there were 276 MSI cases (20%), LS may be considered virtually excluded in the 183 cases which showed positive staining for BRAFV600E, leaving only 93 CRCs (7% of the entire CRC population) requiring formal genetic counseling and further testing.
We note that only selected cases in the TMA cohort underwent genetic testing as part of their clinical care. Therefore this cohort may include some cases of unrecognized LS – a potential limitation of this study. However it is reassuring that all 11 CRCs known to be associated with LS were identified by our IHC approach and that the yield of targeted molecular testing in this BRAF-ve/MSI group is therefore at least 12% (11 of 93).
We caution that all screening strategies have limited sensitivity and specificity. For example, although MMR IHC and formal MSI analysis demonstrate similar sensitivity and specificity, neither will identify all cases of LS.[2, 3] Similarly BRAFV600E mutant CRCs can occur in LS patients, albeit in only a few reported cases (less than 1%).[10-14] These potential pitfalls are well illustrated in our targeted examination of cases from the ACCFR. In this cohort there was one false positive of BRAFV600E IHC compared to allele-specific PCR in a patient with LS and one true positive of BRAFV600E IHC in a tumor from a patient with LS which also harbored this mutation. Therefore regardless of IHC findings we recommend that formal genetic testing be considered in individuals at very high risk for LS based on clinical findings.
BRAF IHC has been shown to be robust and reliable by several different groups studying different tumors. While occasional false negative staining has been reported, false positives (which would result in LS being incorrectly discounted) are rare in experienced hands.[26-32] However, we caution that deployment of BRAFV600E IHC in the clinical setting should be subject to an appropriate quality assurance program with prospective validation and that great care should be taken in optimizing the antibody for use in individual laboratories. We found the antibody performed very well but only after ideal dilutions and antigen retrieval procedures were formulated for our particular laboratory conditions. Although there was commonly non-specific staining of mucus, non-neoplastic epithelium and sometimes smooth muscle cells, non-specific staining was not found in malignant cells once conditions were optimized. During the optimization process or if conditions are not ideal, we would recommend paying careful attention to the degree of non-specific staining in non-neoplastic cells compared to the degree of staining in neoplastic cells before cases which show weak staining are classified as positive or negative. Because BRAFV600E is a cytoplasmic stain, particular care is recommended in interpreting cases with a mucinous histology with minimal cytoplasm.
Identification of the BRAFV600E mutation in MSS tumors may emerge as a beneficial by-product of this approach to universal LS screening in light of studies which demonstrate that these tumors are associated with a significantly worse outcome.[12, 15-19, 21, 22]
We estimate the primary antibody costs of performing BRAFV600E IHC as being less than $10US per case with minimal additional labor costs since MMR IHC is concurrently performed. Of note our study also confirmed the findings of others suggesting that, when interpreted with care by an experienced pathologist, IHC for PMS2 and MSH6 alone identifies all cases of MMR.[40-42] Therefore the cost of BRAFV600E IHC could potentially be completely offset by omitting IHC for MLH1 and MSH2 from LS screening programs.
In conclusion, IHC for BRAFV600E mutation is highly concordant with current PCR-based approaches and effective in the diagnosis of LS. We propose that BRAF and MMR IHC can be performed on all CRC patients at point of care diagnostic testing on surgical excision specimens. One potential algorithm is presented in figure 3. There are clear advantages in terms of laboratory work flow and triaging referral for formal genetic analysis in performing both MMR and BRAF IHC together rather than sequentially and this approach has the added benefit of identifying the emerging poor prognostic group of BRAFV600E-mutated, MSS tumors.
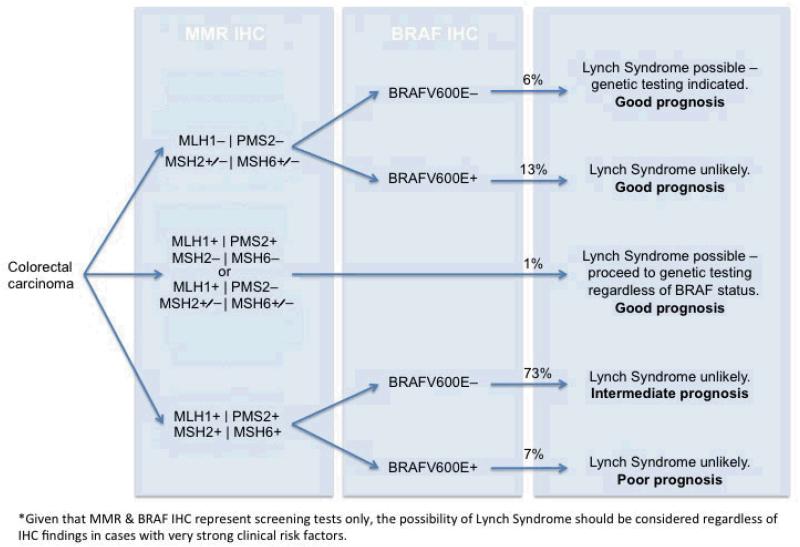
Figure 3.
Algorithm for reflex testing of CRC for both MMR and BRAF by IHC. The percentage incidence of each diagnostic group in our cohort of 1403 colorectal carcinomas is given.
Supplementary Material
Acknowledgements
The authors thank all study participants of the Australasian Colon Cancer Family Registry and Study Co-ordinator Judi Maskiell, Data Managers Kelly Aujard, Maggie Angelakos and David Packenas and laboratory staff Belinda Nagler and Sally-Ann Pearson. The authors also acknowledge the contributions of the late Professor Jeremy Jass to the study including performing pathology reviews for cases.
Funding
National Cancer Institute, National Institutes of Health under RFA #CA-95-011 and through cooperative agreements with members of the Colon-Cancer-Family-Registry and Principal Investigators of Australasian-Colorectal-Cancer-Family Registry (U01CA097735).
Footnotes
Disclosures of potential conflicts of interest: Under a licensing agreement between Ventana Medical Systems, Inc., Tucson, Arizona, and the German Cancer Research Center, DC and AvD are entitled to a share of royalties received by the German Cancer Research Center on the sales of VE1 antibody. The terms of this arrangement are being managed by the German Cancer Research Center in accordance with its conflict of interest policies. SM is a paid consultant for Roche Molecular Advisory Committee. All other authors have no potential conflicts of interest to declare.
Manuscript contribution(s): Conception & design – AG, MW, DB, CT, DC, AvD. Collection & assembly of data – all authors. Data analysis & interpretation – MW, AC, JH, MJ, DB, CT, AG, SM, SC, AdC. Manuscript writing & final approval - all authors. Provision of study material or patients – DC, AvD, CT, AG, MW, DB.
Access to data: Authors CT and AG had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Disclaimer: The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Cancer Family Registries, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the Cancer Family Registry. Authors had full responsibility for the design of the study, the collection of the data, the analysis and interpretation of the data, the decision to submit the manuscript for publication, and the writing of the manuscript.
Ethics approval: Written informed consent was obtained from all study participants who underwent genetic testing. The study protocol was approved by the QIMR HREC under protocol P628 and the Royal North Shore Hospital Ethics Committee under protocol 1201-035M.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Atlanta., A.C.C.F.F.-. American Cancer Society 2011 http://www.cancer.org/research/cancerfactsfigures/colorectalcancerfactsfigures/colorectal-cancer-factsfigures-2011-2013-page.
- 2.Beamer LC, et al. Reflex immunohistochemistry and microsatellite instability testing of colorectal tumors for Lynch syndrome among US cancer programs and follow-up of abnormal results. J Clin Oncol. 2012;30(10):1058–63. doi: 10.1200/JCO.2011.38.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hampel H, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26(35):5783–8. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palomaki GE, et al. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet Med. 2009;11(1):42–65. doi: 10.1097/GIM.0b013e31818fa2db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarvinen HJ, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118(5):829–34. doi: 10.1016/s0016-5085(00)70168-5. [DOI] [PubMed] [Google Scholar]
- 6.Hampel H. Point: justification for Lynch syndrome screening among all patients with newly diagnosed colorectal cancer. J Natl Compr Canc Netw. 2010;8(5):597–601. doi: 10.6004/jnccn.2010.0044. [DOI] [PubMed] [Google Scholar]
- 7.Hall MJ. Counterpoint: implementing population genetic screening for Lynch Syndrome among newly diagnosed colorectal cancer patients--will the ends justify the means? J Natl Compr Canc Netw. 2010;8(5):606–11. doi: 10.6004/jnccn.2010.0045. [DOI] [PubMed] [Google Scholar]
- 8.Teutsch SM, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: methods of the EGAPP Working Group. Genet Med. 2009;11(1):3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kastrinos F, Syngal S. Screening patients with colorectal cancer for Lynch syndrome: what are we waiting for? J Clin Oncol. 2012;30(10):1024–7. doi: 10.1200/JCO.2011.40.7171. [DOI] [PubMed] [Google Scholar]
- 10.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 11.Rajagopalan H, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418(6901):934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 12.Li WQ, et al. BRAF mutations are associated with distinctive clinical, pathological and molecular features of colorectal cancer independently of microsatellite instability status. Mol Cancer. 2006;5:2. doi: 10.1186/1476-4598-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka H, et al. BRAF mutation, CpG island methylator phenotype and microsatellite instability occur more frequently and concordantly in mucinous than non-mucinous colorectal cancer. Int J Cancer. 2006;118(11):2765–71. doi: 10.1002/ijc.21701. [DOI] [PubMed] [Google Scholar]
- 14.Baldus SE, et al. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16(3):790–9. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 15.Kakar S, et al. Clinicopathologic characteristics, CpG island methylator phenotype, and BRAF mutations in microsatellite-stable colorectal cancers without chromosomal instability. Arch Pathol Lab Med. 2008;132(6):958–64. doi: 10.5858/2008-132-958-CCCIMP. [DOI] [PubMed] [Google Scholar]
- 16.Kalady MF, et al. BRAF mutations in colorectal cancer are associated with distinct clinical characteristics and worse prognosis. Dis Colon Rectum. 2012;55(2):128–33. doi: 10.1097/DCR.0b013e31823c08b3. [DOI] [PubMed] [Google Scholar]
- 17.Maestro ML, et al. Role of the BRAF mutations in the microsatellite instability genetic pathway in sporadic colorectal cancer. Ann Surg Oncol. 2007;14(3):1229–36. doi: 10.1245/s10434-006-9111-z. [DOI] [PubMed] [Google Scholar]
- 18.Ogino S, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18(3):890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pai RK, et al. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol. 2012;36(5):744–52. doi: 10.1097/PAS.0b013e31824430d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth AD, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28(3):466–74. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 21.Samowitz WS, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65(14):6063–9. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 22.Zlobec I, et al. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer. 2010;127(2):367–80. doi: 10.1002/ijc.25042. [DOI] [PubMed] [Google Scholar]
- 23.Di Nicolantonio F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26(35):5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 24.NCCN . Clinical Practice Guidelines in Oncology Colon Cancer. 2012. Version 3. [DOI] [PubMed] [Google Scholar]
- 25.Capper D, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122(1):11–9. doi: 10.1007/s00401-011-0841-z. [DOI] [PubMed] [Google Scholar]
- 26.Koperek O, et al. Immunohistochemical Detection of the BRAF V600E-mutated Protein in Papillary Thyroid Carcinoma. Am J Surg Pathol. 2012;36(6):844–50. doi: 10.1097/PAS.0b013e318246b527. [DOI] [PubMed] [Google Scholar]
- 27.Bullock M, et al. Utilisation of a monoclonal antibody for BRAFV600E detection in papillary thyroid carcinoma. Endocr Relat Cancer. 2012;19(6):779–784. doi: 10.1530/ERC-12-0239. [DOI] [PubMed] [Google Scholar]
- 28.Skorokhod A, et al. Detection of BRAF V600E mutations in skin metastases of malignant melanoma by monoclonal antibody VE1. J Am Acad Dermatol. 2012;67(3):488–91. doi: 10.1016/j.jaad.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 29.Preusser M, et al. Expression of BRAF V600E Mutant Protein in Epithelial Ovarian Tumors. Appl Immunohistochem Mol Morphol. 2012 doi: 10.1097/PAI.0b013e31825d7402. [DOI] [PubMed] [Google Scholar]
- 30.Andrulis M, Penzel R, Weichert W, von Deimling A, Capper D. Application of a BRAF V600E Mutation-specific Antibody for the Diagnosis of Hairy Cell Leukemia. Am J Surg Pathol. 2012 doi: 10.1097/PAS.0b013e3182549b50. doi: 10.1097/PAS.0b013e3182549b50. [DOI] [PubMed] [Google Scholar]
- 31.Dietrich S, et al. BRAF inhibition in refractory hairy-cell leukemia. N Engl J Med. 2012;366(21):2038–40. doi: 10.1056/NEJMc1202124. [DOI] [PubMed] [Google Scholar]
- 32.Capper D, et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol. 2012;123(2):223–33. doi: 10.1007/s00401-011-0887-y. [DOI] [PubMed] [Google Scholar]
- 33.Bosman FT, C F, Hruban RH, Theise ND. WHO classification of Tumors of the DIgestive System. 4th ed IARC press; Lyon, France: 2010. [Google Scholar]
- 34.Edge SE, B D, Compton CC. AJCC Cancer Staging Manual. 7th edition Springer; New York, NY, USA: 2009. [Google Scholar]
- 35.James MR, et al. Rapid screening of 4000 individuals for germ-line variations in the BRAF gene. Clin Chem. 2006;52(9):1675–8. doi: 10.1373/clinchem.2006.070169. [DOI] [PubMed] [Google Scholar]
- 36.Buchanan DD, et al. Risk factors for colorectal cancer in patients with multiple serrated polyps: a cross-sectional case series from genetics clinics. PLoS One. 2010;5(7):e11636. doi: 10.1371/journal.pone.0011636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newcomb PA, et al. Colon Cancer Family Registry: An International Resource for Studies of the Genetic Epidemiology of Colon Cancer. Cancer Epidemiology Biomarkers & Prevention. 2007;16(11):2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 38.Walsh MD, et al. Immunohistochemical testing of conventional adenomas for loss of expression of mismatch repair proteins in Lynch syndrome mutation carriers: a case series from the Australasian site of the colon cancer family registry. Mod Pathol. 2012;25(5):722–30. doi: 10.1038/modpathol.2011.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang AH, et al. Optimized allele-specific real-time PCR assays for the detection of common mutations in KRAS and BRAF. J Mol Diagn. 2011;13(1):23–8. doi: 10.1016/j.jmoldx.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shia J, et al. Immunohistochemistry as first-line screening for detecting colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome: a 2-antibody panel may be as predictive as a 4-antibody panel. Am J Surg Pathol. 2009;33(11):1639–45. doi: 10.1097/PAS.0b013e3181b15aa2. [DOI] [PubMed] [Google Scholar]
- 41.Hall G, et al. Immunohistochemistry for PMS2 and MSH6 alone can replace a four antibody panel for mismatch repair deficiency screening in colorectal adenocarcinoma. Pathology. 2010;42(5):409–13. doi: 10.3109/00313025.2010.493871. [DOI] [PubMed] [Google Scholar]
- 42.Mojtahed A, et al. A two-antibody mismatch repair protein immunohistochemistry screening approach for colorectal carcinomas, skin sebaceous tumors, and gynecologic tract carcinomas. Mod Pathol. 2011;24(7):1004–14. doi: 10.1038/modpathol.2011.55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.