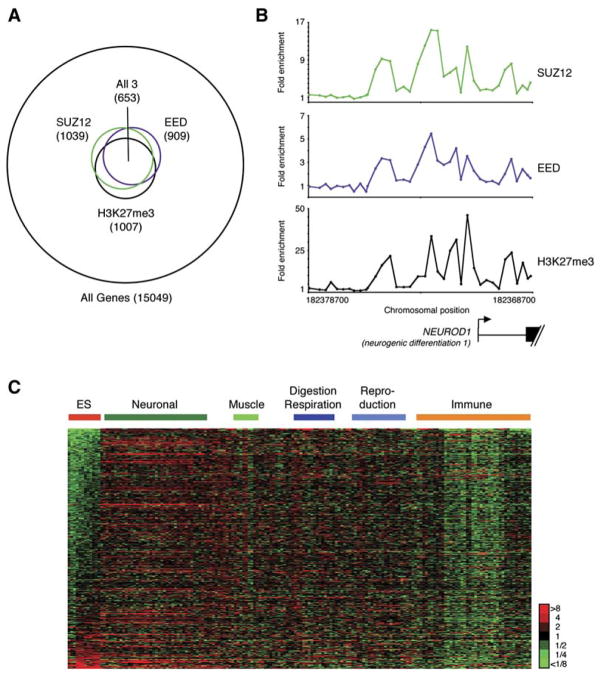
Figure 2. SUZ12 Is Associated with EED, histone H3K27me3 Modification, and Transcriptional Repression in ES Cells.
(A) Venn diagram showing the overlap of genes bound by SUZ12 at high-confidence, genes bound by EED at high-confidence, and genes trimethylated at H3K27 at high-confidence. The data are from promoter micro-arrays that contain probes tiling −8 kb and +2 kb around transcription start. 72% of the genes bound by SUZ12 at high-confidence are also bound by EED at high-confidence; others are bound by EED at lower confidence (Figure S6).
(B) SUZ12 (top), EED (middle), and H3K27me3 (bottom) occupancy at NEUROD1. The plots show unprocessed enrichment ratios for all probes within this genomic region (SUZ12 ChIP versus whole genomic DNA, EED ChIP versus whole genomic DNA, and H3K27me3 ChIP versus total H3 ChIP). Chromosomal positions are from NCBI build 35 of the human genome. NEUROD1 is shown to scale below plots (exons are represented by vertical bars). The start and direction of transcription are noted by arrows.
(C) Relative expression levels of 604 genes occupied by PRC2 and trimethylated at H3K27 in ES cells. Comparisons were made across four ES cell lines and 79 differentiated cell types. Each row corresponds to a single gene that is bound by SUZ12, associated with EED and H3K27me3, and for which Affymetrix expression data are available. Each column corresponds to a single expression microarray. ES cells are in the following order: H1, H9, HSF6, HSF1. For each gene, expression is shown relative to the average expression level of that gene across all samples, with shades of red indicating higher than average expression and green lower than average expression according to the scale on the right. Cell types are grouped by tissue or organ function, and genes are ranked according the significance of their relative level of gene expression in ES cells.