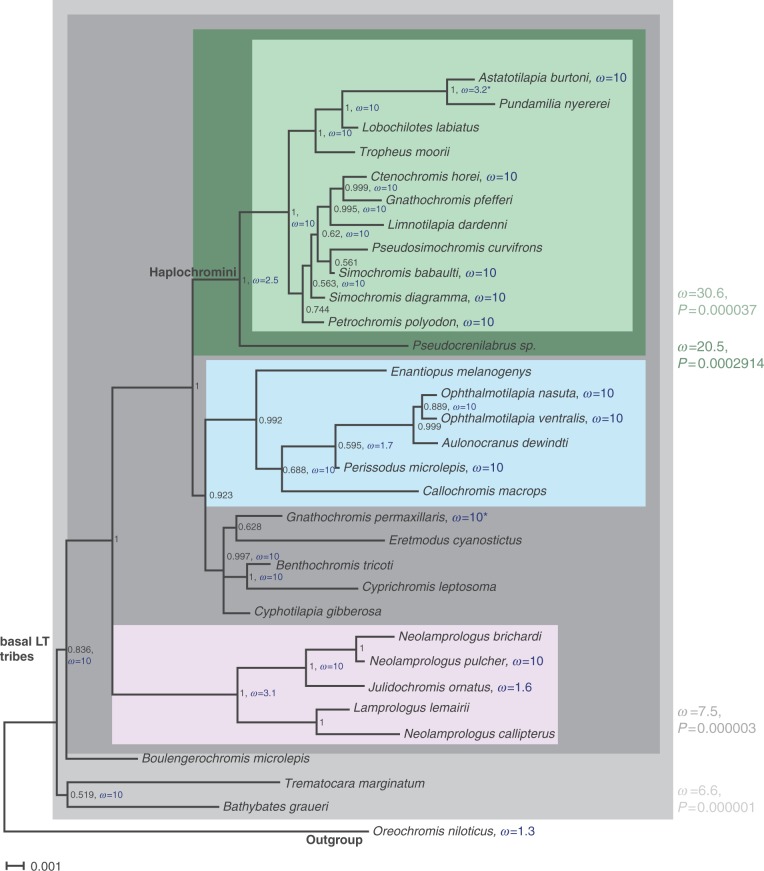
Fig. 2.
Ka/Ks rates on a phylogenetic tree of aromatase cyp19a1A in cichlids. Phylogenetic tree of aromatase cyp19a1A of representatives of the major East African cichlid lineages showing the Ka/Ks rates reconstructed with HyPhy and branch-site selection as calculated with codeml. Nodes are labeled with Bayesian posterior probabilities as obtained with MrBayes. Colored boxes denote clades that were tested against all other branches with codeml (see Materials and Methods for details). Significant obtained Ka/Ks values (ω) are shown next to the boxes in the respective colors. ω values denoted in blue were calculated with Branch-site REL in HyPhy. Table 1 presents the sites that are under positive selection as detected by codeml in the different clades that show a significant ω with a probability (BEB) of more than 50%. The values obtained with Selecton are given in addition.