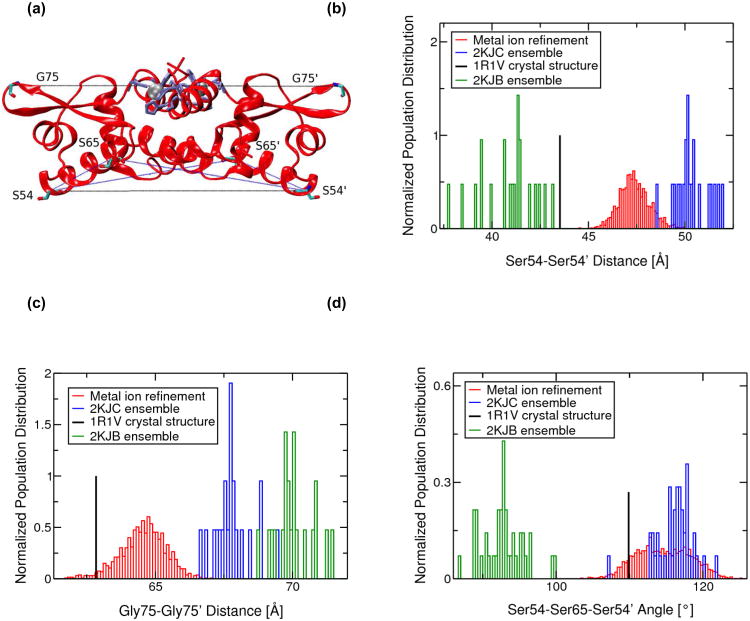
Fig. 6.
(a) Ribbon representation of the MRD-NMR structure of zinc bound CzrA indicating residues used to measure changes between the open and closed conformations. Zinc ions are shown as silver spheres and protein residues are shown in licorice depiction. Residues involved in metal ion binding are colored lilac. Normalized population distributions of (b) Ser54-Ser54' and (c) Gly75-Gly75' inter-subunit Cα-Cα distances and (d) Ser54-Ser65-Ser54' and Ser54'-Ser65'-Ser54 inter-subunit Cα-Cα-Cα angles for the 2KJC ensemble of NMR structures of Zn(II)-CzrA, the 2KJB ensemble of NMR structures of DNA-bound CzrA, and the metal ion refined RDC+NOE constrained MD and QM/MM MD ensemble of Zn(II)-CzrA structures. Distances from the 1R1V Zn(II)-bound crystal structure of CzrA are also indicated