Abstract
In addition to being causally linked to the formation of multiple tumor types, tobacco use has been associated with decreased efficacy of anticancer treatment and reduced survival time. A detailed understanding of the cellular mechanisms that are affected by tobacco smoke should facilitate the development of improved preventive and therapeutic strategies. We have investigated the effects of a tobacco smoke (TS) extract on the transcriptome of MSK-Leuk1 cells, a cellular model of oral leukoplakia. Using Affymetrix HGU133 Plus 2 arrays, 411 differentially expressed probesets were identified. The observed transcriptome changes were grouped according to functional information, and translated into molecular interaction network maps and signaling pathways. Pathways related to cellular proliferation, inflammation, apoptosis and tissue injury appeared to be perturbed. Analysis of networks connecting the affected genes identified specific modulated molecular interactions, hubs and key transcription regulators. Thus, TS was found to induce several EGFR ligands forming an EGFR-centered molecular interaction network, as well as several AhR-dependent genes, including the xenobiotic metabolizing enzymes CYP1A1 and CYP1B1. Notably, the latter findings in vitro are consistent with our parallel finding that levels of CYP1A1 and CYP1B1 were increased in oral mucosa of smokers. Collectively, these results offer insights into the mechanisms underlying the procarcinogenic effects of TS and raise the possibility that inhibitors of EGFR or AhR signaling will prevent or delay the development of tobacco smoke-related tumors. Moreover, the inductive effects of TS on xenobiotic metabolizing enzymes may help explain reduced efficacy of chemotherapy, and suggest targets for chemopreventive agents in smokers.
Keywords: microarray, tobacco smoke, gene expression, oral, aryl hydrocarbon receptor
Introduction
Tobacco use is an important risk factor for multiple human malignancies and accounts for approximately 30% of all cancer-related deaths in the United States (1). Exposure to tobacco has been linked to a variety of malignancies including cancers of the lung, oral cavity and pharynx, esophagus, pancreas, liver, bladder, and cervix (2). More than 100 carcinogens, mutagens and tumor promoters have been identified in tobacco smoke (3, 4). In addition to being a major cause of cancer, smoking can alter the activity of chemopreventive agents (4, 5), stimulate the metabolic clearance of targeted anticancer therapies (6), and increase the risk of second primary tumors (7). Although cessation of tobacco use is highly desirable, it is not realistic for everyone. Hence, there is a significant interest in chemopreventive agents that could protect against the carcinogenic effects of tobacco smoke. Moreover, a clearer understanding of the mechanisms that are modulated by tobacco smoke should lead to more effective treatments resulting in an improved outcome for cancer patients.
Given the significance of tobacco smoke as both a cause of cancer and potential modifier of treatment outcome (1, 8–10), we have investigated the effects of a tobacco smoke (TS) extract on the transcriptome in a cellular model of oral leukoplakia. This model was chosen because the link between exposure to tobacco smoke and head and neck squamous cell carcinoma is well established. Furthermore, smoking reduces the likelihood of treatment response in head and neck cancer patients (11) and increases the risk of second primary tumors in patients who have been successfully treated for their index head and neck malignancy (7). The transcriptome analysis involved the identification of genes differentially expressed due to TS exposure in this cell line, followed by a classification of these genes into domains of putative physiological function. The classification involved the mapping of interactions among differentially expressed genes based on information from interaction databases. Several different databases and tools were employed in this analysis of the observed global transcriptome changes in terms of biological functions and pathways, with the results suggesting that pathways related to cell proliferation, inflammation, apoptosis and tissue injury were affected by TS. Finally, network representations of these data led to identification of proteins in the differentially expressed cohort that have multiple interaction partners (interaction hubs), and transcription factors affected by TS. The analyses identified an epidermal growth factor receptor (EGFR)-centered network comprised of several ligands of the EGFR that were induced by TS. Notably, aryl hydrocarbon receptor (AhR)-dependent genes induced by TS included the enzymes CYP1A1 and CYP1B1, which are of special interest because each may contribute to both carcinogenesis of the aerodigestive tract and drug metabolism (12–14). Consequently, we extended our analysis of TS related transcriptome changes to human volunteers. Consistent with the in vitro findings presented here, we found increased levels of both CYP1A1 and CYP1B1 in the oral mucosa of healthy human subjects who smoked cigarettes. Further comparison of our findings in the MSK-Leuk1 cell model to in vivo data on transcriptome differences in airway epithelial cells of smokers (versus never smokers) (15), identified a canonical set of differentially expressed genes and perturbed pathways. In addition to providing new insights into the procarcinogenic effects of tobacco smoke, these findings highlight the potential of tobacco smoke to alter the efficacy of pharmaceutical agents by inducing the expression of xenobiotic metabolizing enzymes.
Materials and Methods
Materials
Keratinocyte basal and growth media were supplied by Clonetics Corp. (San Diego, CA). MuLV Reverse Transcriptase, Oligo d(T)16 and RNase inhibitor were from Roche Applied Science (Indianapolis, IN), and Taq polymerase from Applied Biosystems (Foster City, CA). HGU133 Plus 2 microarrays were from Affymetrix (Santa Clara, CA).
Tissue culture
The MSK-Leuk1 cell line was established from a dysplastic leukoplakia lesion adjacent to a squamous cell carcinoma of the tongue (16). Cells were routinely maintained in keratinocyte growth medium supplemented with bovine pituitary extract. Cells were grown in basal medium for 24 hr before treatment. Treatment with vehicle (PBS) or TS was carried out on cells grown in growth factor free basal medium. Cellular cytotoxicity was assessed by measurements of cell number, trypan blue exclusion, and release of lactate dehydrogenase. There was no evidence of cytotoxicity in our experiments.
Preparation of tobacco smoke condensate
Cigarettes (2R4F, Kentucky Tobacco Research Institute) were smoked in a Borgwaldt piston-controlled apparatus (model RG-1) using the Federal Trade Commission standard protocol (17). The protocol variables attempt to mimic a standardized human smoking pattern (duration, 2 s/puff; frequency, 1 puff/min; volume, 35 mL/puff). Cigarettes were smoked one at a time in the apparatus and the smoke drawn under sterile conditions into premeasured amounts of sterile PBS (pH 7.4). This smoke in PBS represents whole trapped mainstream smoke abbreviated as TS. Quantitation of smoke content is expressed in puffs/mL with one cigarette yielding about 8 puffs drawn into a 5 mL volume. The final TS concentration in cell culture medium is expressed as puffs/mL medium.
Human tissue
Buccal mucosa specimens were obtained from 9 never smokers and 9 active smokers with a history of at least 10 pack years. Subjects were excluded if they had gross evidence of oral inflammation, a history of heavy alcohol consumption, or recent use of nonsteroidal anti-inflammatory drugs or other anti-inflammatory medications. After topical anesthesia, 5-mm punch biopsies were obtained from grossly normal appearing buccal mucosa. Tissue samples were immediately snap frozen in liquid nitrogen and stored at −80°C until analysis. Hematoxylin and eosin staining of representative formalin-fixed samples indicated that the biopsies were primarily comprised of epithelium. This study was approved by the Committee on Human Rights in Research at Weill Cornell Medical College.
Reverse transcription-PCR
Total cellular RNA was isolated from cells using the RNeasy Mini Kit according to the manufacturer’s instructions. Reverse transcription was performed using 2 μg of RNA per 50 μL of reaction. The reaction mixture contained 1x PCR Buffer II, 2.5 mmol/L MgCl2, 0.5 mmol/L dNTPs, 2.5 μmol/L oligo(dT)16 primer, 50 U RNase inhibitor, and 125 U MuLV. Samples were amplified in a thermocycler for 10 min at 25°C, 42°C for 15 min and 99°C for 5 min, and 5°C for 5 min. The resulting cDNA was then used for amplification. The volume of the PCR reaction was 25 μL and contained 5 μL of cDNA, 1x PCR Buffer II, 2 mmol/L MgCl2, 0.4 mmol/L dNTPs, 400 nmol/L forward primer, 400 nmol/L reverse primer and 2.5 unitsTaq polymerase. Samples were denatured at 95°C for 2 min and then amplified for 30 cycles in a thermocycler under the following conditions: 95°C for 30 s, 62°C for 30 s, and 70°C for 45 s. Subsequently, the extension was carried out at 70°C for 10 min. Primers were synthesized by Sigma Genosys, and the sequences are listed in Supplementary Table 1.
To determine levels of mRNAs for CYP1A1 and CYP1B1 in buccal mucosa, total RNA was isolated from biopsy samples using the RNeasy Mini-kits from Qiagen. Analysis was carried out as described above. Thermal cycling conditions were: 95°C for 2 min, followed by 30 s at 95°C, 30 s at 62°C and 45 s at 72°C for 30 cycles and then 72°C for 10 min. PCR products were subjected to electrophoresis on a 1% agarose gel with 0.5 μg/mL ethidium bromide. The identity of each PCR product was confirmed by DNA sequencing. A computer densitometer (ChemDoc, Bio-Rad) was used to quantify the density of the different bands.
Microarray Procedures
Biotinylated cRNA were prepared according to the standard Affymetrix protocol from 2.5 ug total RNA (http://www.affymetrix.com). Following fragmentation, 10 ug of cRNA were hybridized for 16 hr at 45° C on GeneChip HG U133 Plus 2 arrays. GeneChips were washed and stained in the Affymetrix Fluidics Station 450 and scanned using the Affymetrix GeneChip Scanner 3000. At each of the 5 time points, 6 biological replicates were used for each TS treatment and another 6 biological replicates for vehicle-treated samples. In total, 60 chips were used.
Microarray data analysis
The gene annotations used for each probeset were from the February 2008 NetAffx HGU133 Plus 2 Annotation Files.
Preprocessing
Raw image data were background corrected, normalized and summarized into probeset expression values using the Robust Multichip Average algorithm (RMA). In our analysis, the largest variation in results arose from the method of preprocessing. Both the development of preprocessing methods and the assessment of results are active research areas and the choice of method affects the analysis outcome (18). RMA (19) and a modification, GCRMA (20), have been shown to perform as well as, or better than alternatives using Plasmode data sets. However, GC-RMA can be biased when outliers are not eliminated. Further, both GCRMA, and another commonly used method, Affymetrix Microarray Analysis Suite v5.0 (MAS5), may perform poorly with highly variable human data (21). To check for the robustness of our results to different preprocessing methods, raw image data were preprocessed using both the RMA algorithm (19, 22) within GeneSpring 7.2 Software (Agilent Technologies), and MAS5, and then analyzed statistically as described below. We found a 75% agreement between RMA-preprocessed results and those from MAS5 (data not shown). This indicates a high level of consistency, as the overlap between RMA and MAS cited in the literature ranges from 27% (23) to 70% (18). In light of evaluation of the currently applicable methods in recent reviews (24), we performed both the statistical and functional inference analysis from the RMA-preprocessed data.
Normalization and Filtering
The data from each chip were normalized for inter-array comparisons as follows: measurements of <0.01 were set to 0.01 and each chip was normalized to 50% of the measurements taken from that chip (a procedure considered appropriate for large arrays when most of the genes are unaffected by experimental parameters). We further applied a filter to remove probesets that were not reliably detected. From the complete set of ~54675 probesets on the HGU133 Plus 2 array, for every time point, we filtered out probesets whose minimum raw expression level was not 50 in at least 2 out of 12 conditions. This cutoff was chosen from the scatter plot distribution of expression values for TS vs. vehicle-treated controls (marked as C). We further filtered out probesets with low confidence if their t-test p-value was not <0.05 in at least 2 out of 12 conditions, using the Benjamini and Hochberg false discovery rate criterion (25). Genes that passed these tests were defined as expressed and were statistically analyzed. This set of analyzed genes consists of the following numbers of probesets: 20,791 (at 0.5 hr), 20,443 (3 hr), 20,979 (6 hr), 19,584 (12 hr) and 20,034 (24 hr).
Statistical Analysis
Probesets were analyzed using both ANOVA and Significance Analysis of Microarrays (SAM). Genes that passed both SAM and ANOVA tests and had normalized expression values altered by a factor of 1.5 fold were deemed significant. ANOVA analysis was performed using GeneSpring 7.2 software. SAM analysis was performed using SAM Microsoft Excel plug-in. The details of this analysis are as follows: A 1-way ANOVA was performed at every time point using parametric test, not assuming variances equal (Welch’s t-test) with p-value <0.05. To address the problem of multiple comparisons, Benjamini and Hochberg multiple testing correction was employed to maintain False Discovery Rate (FDR) at 5% (25). The SAM method utilizes a set of gene-specific t-tests, where each gene is assigned a score on the basis of its change in gene expression, relative to the standard deviation of repeated measurements for that gene (26). Genes with scores greater than threshold delta values at FDR ~4–5% were deemed significant (as the FDR is dependent on delta value, an exact 5% FDR is not possible using the delta-slider within the SAM Excel plug in). At this delta, the fold change parameter was set to 1.5. For input to SAM, two-class unpaired response was chosen on normalized data with t-statistics, 200 permutations and default SAM options. In summary, the analysis steps described above in comparing TS vs C at every time point are: Preprocessing (RMA) → Normalization → Filtering for Expression → Statistical Analysis (SAM and ANOVA with multiple testing correction) → Filtering for Fold Change.
Comparative Analysis
Raw data (Affymetrix HGU-133A chips) were downloaded from GEO repository (http://www.ncbi.nlm.nih.gov/geo/). Smoking status related genes in airway epithelia that are significantly differentially expressed were assessed using GeneSpring 7.2 as described above and separated into upregulated and downregulated gene sets comprising 110 and 21 genes, respectively. Gene Set Enrichment Analysis (GSEA) (27) was used to compare the MSK-Leuk1 and airway epithelia gene expression results. The up- and downregulated gene sets in airway data were used to identify enrichment in the MSK-Leuk1 data. The MSK-Leuk1 expression data inputs were the normalized ratio values of all probesets in HGU133 Plus 2 arrays and their collapsed HUGO symbols. Gene set enrichment analysis was performed for every time point using default parameters, except the permutation type, which was gene set instead of phenotype (as recommended by the GSEA manual when number of replicates are <7). A reverse analysis was also performed by using all the probesets in airway epithelial dataset as input, and searching for enrichment on the significantly up- or downregulated gene sets at every time point of MSK-Leuk1 cells exposed to TS. Here, the differentially expressed gene sets in MSK-Leuk1 data were collapsed to the HUGO symbols of probesets available in HGU-133A microarrays.
Leading-Edge Subset
The leading-edge subset is defined as comprising those members of the gene set that appear in the ranked list at, or before, the point where the running sum reaches its maximum deviation from zero. The set is suggested to be the core of a gene set that accounts for the enrichment score (27).
Functional Analysis
To relate the results to cell physiological mechanisms, the transcriptional data were integrated with available experimental signaling data for TS. The complex biological processes induced by TS were examined in the context of detailed protein-protein interaction maps (28), and molecular networks (29). The interaction networks shown in Fig. 1C and 2A were generated with Ingenuity Pathways Analysis (IPA), a web-delivered application used to discover, visualize and explore relevant networks (29). Affymetrix probe identifiers were uploaded to IPA, each identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledgebase and only direct interactions were considered. For the network in Fig. 2A, interactions were queried between these gene objects and all other gene objects stored within IPA to generate a set of networks that were then merged. The only putative hubs considered in the merged network were transcription regulators that are expressed in MSK-Leuk1 cells and have at least 6 direct interactions with differentially expressed genes.
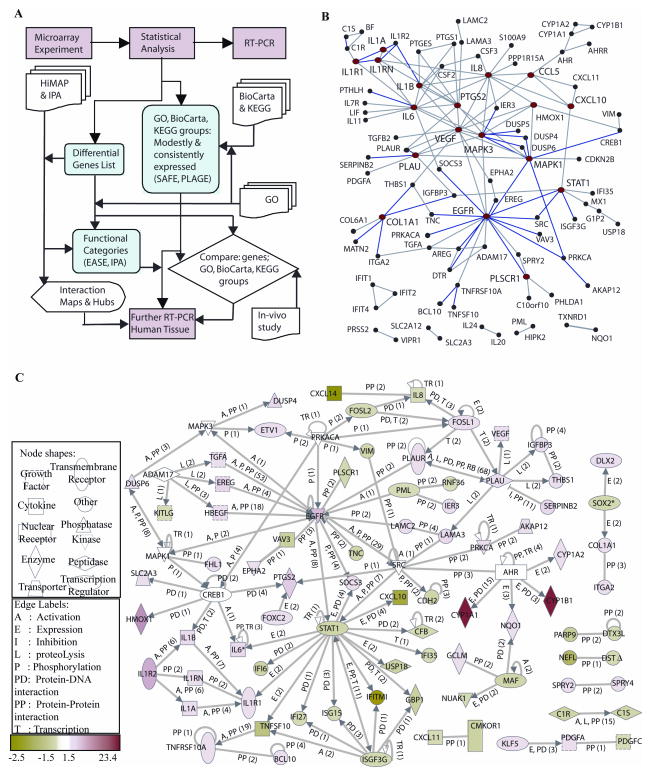
Figure 1.
A, Flowsheet describing the systematic analysis of TS induced changes in the transcriptome of MSK-Leuk1 cells, including hypothesis generation steps.
B, Protein-protein interaction network of differentially expressed genes (www.physiology.med.cornell.edu/go/smoke) and signaling proteins known to be activated by TS (AhR, ADAM17, CREB1, MAPK3 (ERK1), MAPK1 (ERK2), PRKACA and SRC) within HiMAP database, which includes interactions that are literature-confirmed from the Human Protein Reference Database (40) (blue), yeast two-hybrid-defined (gray), or predicted based on function (gray).
Hubs with 4 or more connections (red) are CCL5, COL1A1, CXCL10, EGFR, HMOX1, IL1A, IL1B, IL1RN, IL1R1, IL6, IL8, MAPK1, MAPK3, PTGS2, PLAU, PLSCR1, STAT1 are VEGF. C, Direct interaction network of differentially expressed genes and known TS-activated signaling proteins generated using IPA (29). At each edge, the interaction type is shown and the number of publications is indicated in parentheses. Hubs are listed in Supplementary Table 2.
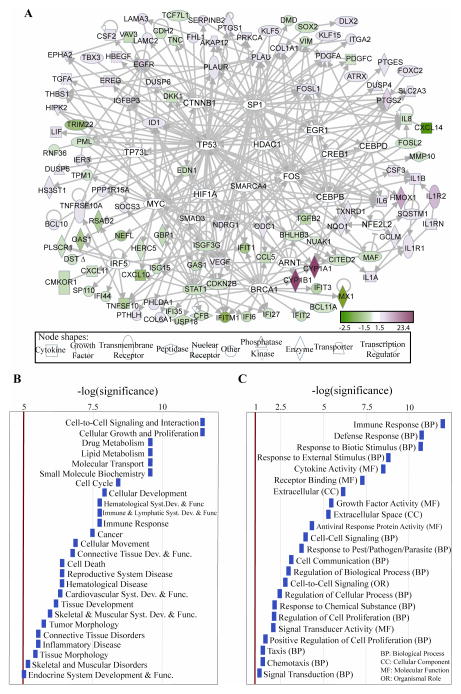
Figure 2.
A, Direct interaction network of differentially expressed genes in TS-treated MSK-Leuk1 cells, integrated with genes related to signaling proteins known to be activated by TS. The white nodes are genes with no significant expression change due to TS in our analysis, but which are expressed in MSK-Leuk1 cells (filtered as in methods, statistical analysis 2a,b) and interact with at least 6 differentially expressed genes, suggesting potential roles for these in TS perturbed signaling processes. These include ARNT, BRCA1, CEBPB, CEBPD, CREB1, CTNNB1, EGR1, FOS, HDAC1, HIF1A, IRF5, MYC, NFE2L2, SMAD3, SMARCA4, SP1, TP53, TP73L. IPA tool (29) was used to generate the panel A. B, Statistically significant diseases and physiological, cellular or molecular functions of the differentially expressed genes. The bars are sized according to the calculated -log (significance) score. C, Statistically overrepresented groups in GO Biological Process (GO BP), GO Molecular Function (GO MF), GO Cellular Components (GO CC) and organismal role, among the differentially expressed genes as compared to the set of genes in the microarray, generated with EASE software, based on overrepresented chromosomes, organismal roles, GO categories, and pathway categories within KEGG, GenMAPP (http://www.genmapp.org), and BBID (http://bbid.grc.nia.nih.gov) databases. Categories with Bonferroni p-value<0.05 were assumed significant, and Affymetrix identifiers are used.
For functional categories, Ingenuity Knowledgebase (29) and Gene Ontology (GO) (http://www.geneontology.org) databases were searched for categories statistically enriched in the differentially expressed genes set, and the likelihood of perturbations in each category was scored. In searching GO categories, the EASE software (30) was used to compare the Affymetrix probe identifiers of differentially expressed genes with the list of all probes in the HGU133 Plus 2 microarray.
We reasoned that a small but coherent difference in the expression of a group of genes in a category or pathway can be more important than large differences in unrelated genes. Therefore, we carried out categorization and pathway-enrichment analyses that enabled identification and scoring of both significant and modest perturbations in corresponding gene groups. For a functional analysis that can capture modest perturbations in functional groups, we used all expressed genes (as defined above in the Microarray data analysis section). Hence, we identified groups of genes that correspond to specific enzymatic, metabolic or signaling pathways within pathway databases of KEGG (http://www.genome.jp/kegg), and BioCarta (http://biocarta.com/genes/index.asp) using the PLAGE tool (31) and the SAFE tool (32) within GO categories. Note that we did not employ an FDR or FWER error estimate for functional groups. While this might result in an increase in Type II estimates, it is acceptable because we focused particularly on pathways that appear to be consistently perturbed at multiple time points.
Clustering
An unsupervised hierarchical clustering analysis across all samples of the microarray data was performed for the probesets found to be differentially expressed between control and TS treated cells at any of the 5 time points (using log-transformed normalized data). A Pearson correlation (uncentered) similarity metric and average linkage clustering was performed with CLUSTER and TREEVIEW software obtained at http://rana.lbl.gov/EisenSoftware.htm (see Supplementary Figure 1). Our study was not designed to identify dynamic effects of TS so that limitation to 5 time points results in a rather sparse matrix for such analysis under the commonly utilized tools (CAGED (33) and SSCLUST (34)). Further results from the use of a newly described clustering algorithm, EP_GOS_Clust (35) were used to classify the time-dependent patterns of changes in gene expression, and will be reported as a separate study.
Additional Information
The complete list of differentially expressed genes and functional groups at every time point, is made available through an interactive web site established as a resource of the Institute for Computational Biomedicine at http://physiology.med.cornell.edu/go/smoke. Also available at that site are the results from the functional analyses of the invivo data, for comparative purposes. The microarray data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the GEO Series accession no. GSE10063.
Results
The effect of TS on gene expression was determined in MSK-Leuk1 cells. Based on microarray analysis, exposure to TS led to at least a 1.5-fold change in the expression of 411 probesets. The complete list of genes, and the expression levels measured under the experimental conditions described here, are available at http://physiology.med.cornell.edu/go/smoke. The number of differentially expressed genes was observed to increase with duration of exposure to TS, amounting to 91, 104, 106, 166 and 274 probesets at 0.5 hr, 3 hr, 6 hr, 12 hr and 24 hr, respectively. The heatmap representation of the results from the clustering of the differentially expressed genes (see Methods), are shown in Supplementary Figure 1. The results show that replicate samples at each time point cluster together and that TS-treated samples and controls are separated into distinct clusters. 27 probesets corresponding to 20 unique genes were differentially expressed at every time point signifying a persistent change in expression (Table 1). Subsequently, RT-PCR was used to validate microarray findings for a subset of 10 differentially expressed genes. The observed changes in expression were quantitatively consistent with the microarray results in showing that exposure to TS led to increased levels of mRNAs for CYP1A1, CYP1B1, PTHLH, IL1B, EREG, ALDH1A3, IL6 and IL24, and to reduced expression of TNFSF10 and CCL5 (data not shown). Thus, the RT-PCR results were entirely consistent with the microarray predictions for all 10 genes evaluated, validating the experiment and statistical analyses. Furthermore, our findings are consistent with published studies in which treatment of MSK-Leuk1 cells with TS induced the following genes: cyclooygenase COX-2 (PTGS2) (17), amphiregulin (AREG) (17, 36) and transforming growth factor alpha (TGFA) (17).
Table 1.
Time-dependent effects of TS on gene expression in MSK-Leuk1 cells. Fold-differences and P values are shown at each time point. Detailed lists and annotations are provided at http://physiology.med.cornell.edu/go/smoke.
| Gene Name | Affy ID | 0.5 hr | 3 hr | 6 hr | 12 hr | 24 hr | Description | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fold | P | fold | P | Fold | P | fold | P | fold | P | |||
| CYP1A1 | 205749_at | 13.4 | 3.3E-06 | 17.4 | 2.1E-08 | 23.4 | 1.1E-07 | 11.3 | 1.5E-06 | 14.3 | 1.0E-05 | cytochrome P450, family 1, subfamily A, polypeptide 1 |
| CYP1B1 | 202437_s_at | 8.3 | 4.2E-06 | 14.1 | 4.0E-06 | 13.5 | 1.7E-06 | 5.8 | 5.5E-06 | 6.1 | 2.1E-08 | cytochrome P450, family 1, subfamily B, polypeptide 1 |
| CYP1B1 | 202435_s_at | 8.1 | 3.8E-06 | 11.4 | 1.2E-06 | 12.1 | 1.7E-06 | 6.7 | 4.3E-05 | 6.1 | 1.2E-04 | cytochrome P450, family 1, subfamily B, polypeptide 1 |
| CYP1B1 | 202436_s_at | 7.4 | 4.3E-06 | 11.0 | 4.5E-05 | 11.2 | 5.5E-08 | 6.0 | 1.5E-06 | 5.9 | 1.7E-04 | cytochrome P450, family 1, subfamily B, polypeptide 1 |
| IL24 | 206569_at | 2.8 | 5.8E-04 | 3.4 | 4.0E-06 | 2.8 | 3.6E-04 | 3.7 | 7.9E-04 | 2.8 | 1.7E-03 | Interleukin 24 |
| PTGS2 | 1554997_a_at | 2.7 | 7.0E-03 | 3.0 | 1.0E-03 | 3.0 | 4.0E-03 | 3.1 | 2.3E-03 | 3.0 | 4.8E-04 | prostaglandin-endoperoxide synthase 2 (COX-2) |
| EREG | 205767_at | 2.1 | 1.8E-03 | 2.6 | 4.0E-06 | 2.2 | 3.8E-05 | 2.4 | 8.2E-05 | 2.0 | 1.0E-04 | Epiregulin |
| LOC151438 | 1560679_at | 2.1 | 8.7E-03 | 1.9 | 1.4E-02 | 1.5 | 3.7E-02 | 3.2 | 2.1E-02 | 5.9 | 6.8E-05 | hypothetical protein LOC151438 |
| PTGS2 | 204748_at | 2.1 | 1.2E-04 | 2.7 | 1.6E-04 | 2.4 | 2.6E-03 | 3.2 | 6.1E-04 | 2.9 | 1.5E-04 | prostaglandin-endoperoxide synthase 2 (COX-2) |
| TIPARP | 212665_at | 2.0 | 2.2E-06 | 3.0 | 9.3E-09 | 2.2 | 1.7E-06 | 2.2 | 1.5E-06 | 2.0 | 1.0E-04 | TCDD-inducible poly (ADP-ribose) polymerase |
| IL1B | 205067_at | 1.9 | 1.3E-04 | 2.2 | 3.5E-07 | 2.1 | 3.1E-06 | 2.1 | 5.5E-06 | 2.1 | 6.4E-05 | Interleukin 1, beta |
| ALDH1A3 | 203180_at | 1.9 | 1.9E-04 | 2.0 | 9.8E-09 | 2.4 | 2.8E-06 | 2.0 | 1.3E-03 | 2.8 | 2.8E-04 | Aldehyde dehydrogenase 1 family, member A3 |
| IL1R2 | 211372_s_at | 1.9 | 8.4E-06 | 1.7 | 1.9E-02 | 1.9 | 5.6E-04 | 2.9 | 2.2E-03 | 5.0 | 4.6E-04 | Interleukin 1 receptor, type II |
| IL1R2 | 205403_at | 1.8 | 1.1E-05 | 1.6 | 1.6E-02 | 1.8 | 5.2E-03 | 2.7 | 3.8E-03 | 5.3 | 6.6E-04 | Interleukin 1 receptor, type II |
| IL1B | 39402_at | 1.8 | 2.9E-05 | 2.1 | 2.1E-08 | 2.0 | 1.7E-06 | 1.9 | 5.5E-06 | 2.2 | 9.4E-05 | Interleukin 1, beta |
| IL20 | 224071_at | 1.7 | 8.3E-03 | 2.1 | 6.4E-05 | 1.8 | 4.6E-04 | 1.7 | 1.5E-02 | 1.7 | 2.7E-03 | Interleukin 20 |
| DUSP4 | 204014_at | 1.7 | 1.3E-02 | 1.6 | 2.5E-03 | 1.7 | 1.2E-04 | 2.3 | 2.3E-04 | 2.4 | 5.4E-04 | dual specificity phosphatase 4 |
| 226034_at | 1.6 | 8.6E-03 | 1.6 | 2.6E-04 | 1.5 | 3.4E-04 | 2.1 | 2.4E-03 | 2.0 | 4.4E-04 | Clone IMAGE:3881549, mRNA | |
| SERPINB2 | 204614_at | 1.6 | 2.9E-03 | 1.6 | 5.8E-04 | 1.6 | 2.4E-04 | 1.9 | 1.8E-04 | 2.5 | 1.0E-05 | serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 2 |
| LOC344887 | 241418_at | 1.5 | 5.0E-02 | 3.6 | 2.9E-05 | 4.2 | 5.5E-08 | 2.6 | 8.8E-05 | 1.9 | 9.8E-04 | Similar to NmrA-like family domain containing 1 |
| MAF | 209348_s_at | −1.5 | 1.9E-04 | −1.5 | 2.7E-02 | −1.5 | 1.8E-02 | −1.7 | 2.1E-02 | −1.8 | 1.7E-03 | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) |
| PRICKLE1 | 226069_at | −1.6 | 4.1E-02 | −1.6 | 1.2E-02 | −2.2 | 2.0E-03 | −1.6 | 2.8E-02 | −2.0 | 7.7E-04 | Prickle homolog 1 (Drosophila) |
| NAV2 | 218330_s_at | −1.7 | 3.8E-03 | −1.8 | 1.3E-02 | −1.7 | 2.0E-03 | −1.7 | 3.7E-03 | −1.8 | 1.0E-04 | Neuron navigator 2 |
| FILIP1L | 204135_at | −1.7 | 3.8E-06 | −2.1 | 7.1E-06 | −2.2 | 1.3E-05 | −2.4 | 1.9E-03 | −2.1 | 6.4E-05 | Filamin A interacting protein 1-like |
| FILIP1L | 1554966_a_at | −1.7 | 8.4E-06 | −2.2 | 6.4E-05 | −2.2 | 2.0E-06 | −2.5 | 1.5E-03 | −2.2 | 1.0E-04 | Filamin A interactiong protein 1-like |
| C10orf10 | 209183_s_at | −2.0 | 6.1E-04 | −2.3 | 1.2E-03 | −2.1 | 4.4E-05 | −2.3 | 4.3E-05 | −2.1 | 6.4E-05 | chromosome 10 open reading frame 10 |
| TNFSF10 | 202688_at | −2.0 | 8.7E-03 | −1.7 | 1.3E-02 | −2.4 | 4.2E-03 | −2.3 | 4.8E-04 | −2.4 | 1.9E-04 | tumor necrosis factor (ligand) superfamily, member 10 |
Interpreting the global transcriptome changes in terms of biological functions and pathways
Several databases and tools were used to classify the differentially expressed genes into relevant molecular, physiological and disease categories. TS induced transcriptome changes were related to cellular processes and pathways using molecular network maps, hubs, functional classes and enrichment analysis. Details of this rigorous functional analysis are represented in a flowsheet format in Figure 1A.
1. Network Analysis
The differentially expressed gene set was integrated with the set of signaling proteins previously shown to be activated by TS: ADAM17 (37), AhR (36, 38), cAMP responsive element binding protein 1 [CREB1] (36), mitogen activated protein kinase (ERK1/2) (39), protein kinase A catalytic subunit [PRKACA] (36) and SRC (unpublished data). Assuming that highly interconnected networks likely represent significant biological function, we sought to identify the interrelations among these genes.
1.1. Protein-Protein Interactions
Direct physical relationships between proteins associated with the gene set were identified utilizing the human protein-protein interaction map, HiMAP (28), as shown in Fig 1B. The proteins found to have 4 or more possible physical interactions are listed in Supplementary Table 2. Hubs with the most Human Protein Reference Database (HPRD) (40) connections (blue edges in Fig. 1B) are MAPK3, COL1A1 and EGFR. In fact, an EGFR-centered subnetwork was observed, where EGFR and its ligands EREG, AREG, DTR (HB-EGF) and TGFA are all significantly induced.
1.2. Interactions based on mammalian biology data
The interactions within the set of TS-modified genes were mapped in the context of the network of physical, transcriptional and enzymatic interactions observed in mammals. The results (obtained using the Ingenuity Pathway Analysis Tool, IPA (29)) are represented in Fig 1C. The interactions considered were: proteolysis, inhibition, protein-protein interaction, expression, protein-DNA interaction, activation and transcription (29). Among hub proteins (listed in Supplementary Table 2), EGFR, PLAU, IL1R1 and related proteins are mainly induced (shown in red), while STAT1 and related proteins are repressed (shown in green). Interestingly, cytokines form a subnetwork composed of IL1R1, IL1R2, IL1A, IL1B and IL1RN, suggesting a TS-induced inflammatory process. Another hub, formed around PLAU, involves genes that contribute to invasiveness (39). At the stringency level used for the initial analysis of TS-affected gene expression, AhR expression was not considered significantly altered, but the likely role of the AhR is evident from the effect of TS on genes known to be regulated by this receptor. Specific examples include the observed increased levels of CYP1A1, CYP1B1 and AREG, which are known targets of ligand-activated AhR (36, 38). In a similar manner, various transcription regulators connected to large numbers of differentially expressed genes can have important roles in signaling pathways perturbed by TS. Mapping the direct interaction network of differentially expressed genes and transcription regulators can reveal such involvement. Shown in Fig. 2A are highly connected putative transcription regulators in response to TS, including sp1 transcription factor [SP1], catenin beta 1 [CTNNB1], interferon regulatory factor 5 [IRF5], histone deacetylase 1 [HDAC1], tumor protein p53 [TP53], tumor protein p73-like [TP73L], hypoxia inducible factor 1, alpha subunit [HIF1A], nuclear factor (erythroid-derived 2)-like 2 [NFE2L2], cellular oncogene c-fos [FOS], myc proto-oncogene protein [MYC], early growth response [EGR1], and a number of well studied transcription factors and cancer-related genes that included [BRCA1], [CEBPB], [CEBPD], and the AhR nuclear translocator [ARNT]. We note that AhR and hypoxia signaling pathways have been demonstrated to crosstalk via the involvement of HIF1α (41), and while HIF1α does not appear to be significantly altered, our analysis suggests its involvement in signaling pathways as a transcription regulator hub.
2. Gene Classification
2.1. Classification of Statistically Significant Genes into Biological Categories
The genes affected by TS were also classified into related diseases and molecular, cellular and physiological functions based on the information from the largest literature curated information database, IPA (Figure 2B) (29). Cancer is the most relevant disease and the most relevant functions are cell-to-cell signaling and interaction, cellular growth and proliferation, drug and lipid metabolism, immune response and connective tissue development and function. We further categorized genes in terms of relevant functional categories using public databases of controlled vocabulary including Gene Ontology (Figure 2C, Supplementary Table 3). The information from these databases also confirms that genes related to cell proliferation and immune responses are overrepresented. As the examples above suggest, the classification of differentially expressed genes is inherently sensitive to the statistical analysis results and filtering criteria. We therefore identified an additional category of statistically significant modestly perturbed functional groups of genes among all expressed genes in the MSK-Leuk1 cells, and defined their functional categories using the GO, KEGG, and BioCarta databases. Pathways consistently perturbed at multiple time points are given in Table 2.
Table 2.
Treatment of MSK-Leuk1 cells with TS affected pathways related to cell proliferation, inflammation, apoptosis, coagulation and tumor suppression. P values are presented. Complete lists per time point are provided at http://physiology.med.cornell.edu/go/smoke. Generated utilizing PLAGE tool (31).
| 0.5 hr | 3 hr | 6 hr | 12 hr | 24 hr | |
|---|---|---|---|---|---|
| KEGG PATHWAYS | P(genes/path) | P(genes/path) | P(genes/path) | P(genes/path) | P(genes/path) |
| Cytokine-cytokine receptor interaction | ↑0.0025(84/257) | ↑<1E-4(86/257) | ↑9E-04(83/257) | ↑0.0008(79/257) | ↑0.0025(83/257) |
| Prostaglandin and leukotriene metabolism | ↑0.0447(15/39) | ↓0.007(15/39) | ↑0.044(15/39) | ↑0.008(15/39) | ↑0.0025(15/39) |
| Neuroactive ligand-receptor interaction | ↑0.0025(43/295) | ↓0.032(46/295) | ↓0.02(51/295) | ↑0.0025(39/295) | ↑<1E-4(46/295) |
| gamma-Hexachlorocyclohexane degradation | ↑0.0092(13/45) | ↑0.003(13/45) | ↑0.008(13/45) | ↑0.0025(11/45) | ↑0.0041(14/45) |
| Hedgehog signaling pathway | ↑0.0117(21/53) | ↑0.003(21/53) | ↓0.009(23/53) | ↑0.0025(20/53) | ↓0.0025(22/53) |
| Complement and coagulation cascades | ↑0.0025(20/69) | ↑0.003(21/69) | ↑0.008(21/69) | ↑0.0025(18/69) | ↑0.0025(19/69) |
| Porphyrin and chlorophyll metabolism | ↓0.003(16/32) | ↑0.011(16/32) | ↑0.0132(16/32) | ↑0.0203(16/32) | |
| Aminosugars metabolism | ↑0.0025(24/27) | ↓0.007(24/27) | ↑0.0349(24/27) | ↑0.0218(24/27) | |
| Jak-STAT signaling pathway | ↑0.0025(81/159) | ↑0.012(81/159) | ↑0.0025(78/159) | ↑0.0025(81/159) | |
| BIOCARTA PATHWAYS | |||||
| Msp/Ron Receptor Signaling Pathway | ↓0.0001(4/6) | ↓0.0025(5/6) | ↓0.0134(5/6) | ↓0.01(5/6) | ↓0.0025(5/6) |
| Cytokine Network | ↑0.0025(5/22) | ↑0.0025(5/22) | ↑0.0025(5/22) | ↑0.003(5/22) | ↑0.0025(5/22) |
| Regulation of hematopoiesis by cytokines | ↑0.0025(6/15) | ↑0.0025(6/15) | ↑0.0418(6/15) | ↑0.046(6/15) | ↑0.0025(6/15) |
| Cytokines and Inflammatory Response | ↑0.0025(11/29) | ↑<1E-4(11/29) | ↑0.0008(11/29) | ↑0.003(11/29 | ↑<1E-4(11/29) |
| Fibrinolysis Pathway | ↑0.0025(6/12) | ↑0.0049(6/12) | ↑0.0025(6/12) | ↑0.003(5/12) | ↑0.0044(6/12) |
| Mechanism of Acetaminophen Activity and Toxicity | ↑0.0025(3/6) | ↑0.0025(3/6) | ↑0.0025(3/6) | ↑<1E-4(3/6) | ↑0.0025(3/6) |
| Signal transduction through IL1R | ↑0.0152(28/33) | ↑0.0025(28/33 | ↑0.0095(28/33) | ↑0.003(28/33) | ↑0.0044(28/33) |
| Erythrocyte Differentiation Pathway | ↑0.0025(7/15) | ↑0.0025(7/15) | ↑0.0025(7/15) | ↑0.013(7/15) | ↑0.0044(7/15) |
| Mechanism of Gene Regulation by Peroxisome Proliferators via PPARa | ↑0.0025(43/57) | ↑0.0197(42/57) | ↑0.013(42/57) | ↑0.0096(42/57) | |
| Cells and Molecules involved in local acute inflammatory response | ↑0.0025(7/17) | ↑0.0364(8/17) | ↓0.0169(8/17) | ↑0.0025(7/17) | |
| IL-10 Anti-inflammatory Signaling Pathway | ↑0.0025(11/13) | ↓0.0076(11/13) | ↑0.0025(11/13) | ↓0.0025(11/13) | |
| Stress Induction of HSP Regulation | ↑0.0025(12/15) | ↑0.0025(12/15) | ↑0.006(11/15) | ↓0.0044(11/15) | |
| Extrinsic Prothrombin Activation Pathway | ↑0.0393(5/13) | ↑0.0291(4/13) | ↑0.013(4/13) | ↑0.0189(4/13) | |
| p53 Signaling Pathway | ↓0.0076(13/15) | ↓0.0446(14/15) | ↓0.013(13/15 | ↓0.0025(13/15) | |
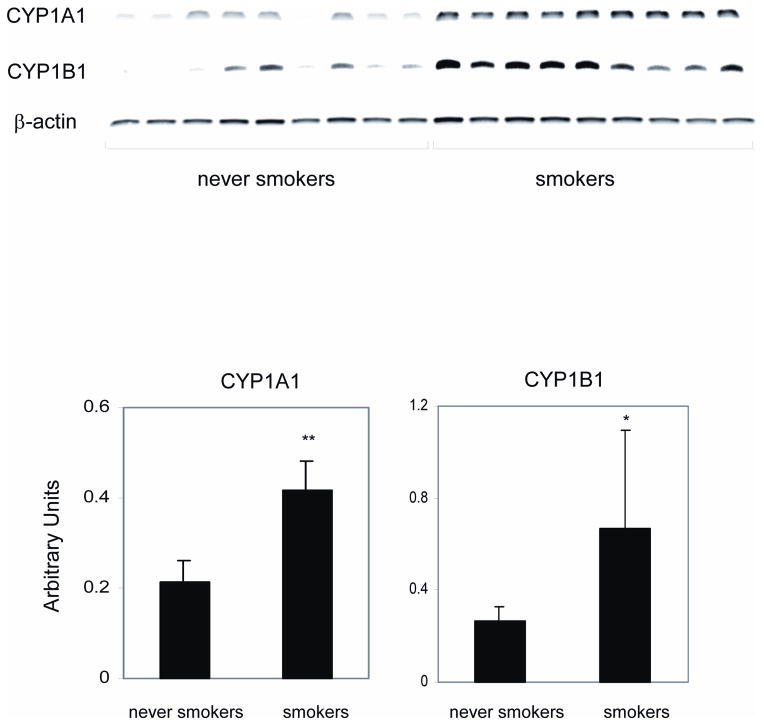
Increased levels of CYP1A1 and CYP1B1 were detected in the oral mucosa of human cigarette smokers
Because the analysis suggested that CYP1A1 and CYP1B1, AhR-dependent genes, were the two genes most induced by TS treatment of MSK-Leuk1 cells (Table 1), we next investigated whether the in vitro results were predictive of increased levels of CYP1A1 and CYP1B1 mRNAs in the oral mucosa of human smokers. To this end, we carried out a comparative analysis in the oral mucosa of healthy cigarette smokers versus never smoking human volunteers. Consistent with the findings in MSK-Leuk1 cells (Table 1), amounts of CYP1A1 and CYP1B1 mRNAs were both increased in the oral mucosa of smokers (Fig. 3).
Figure 3.
Levels of CYP1A1 and CYP1B1 are increased in the oral mucosa of cigarette smokers. A, oral mucosal biopsies were obtained from both never smoking (never smokers, n=9) and smoking (smoker, n=9) human volunteers. Total cellular RNA was extracted from the oral mucosal biopsy samples and reverse transcribed. The expression of CYP1A1 and CYP1B1 mRNAs was assessed by RT-PCR. No bands were observed when cDNA was omitted from the PCR reaction or when the reverse transcriptase enzyme was not included in the reverse transcriptase reaction. B, Results of the data shown in panel A expressed in arbitrary units. Means and S.D. are shown; *P <0.05, **P<0.01 vs. never smokers.
Comparison to airway epithelial transcriptome of smokers and nonsmokers
The acute effects of TS on the transcriptome of MSK-Leuk1 cells were compared to published chronic transcriptome differences measured in airway epithelial cells of 34 current smokers and 23 never smokers (15). To minimize preprocessing and statistical methodology-based differences, these human data were reanalyzed using the same statistical tools employed here. The comparison revealed that the genes strongly overexpressed due to TS in MSK-Leuk1 cells, namely CYP1A1, CYP1B1, ALDH1A3, GCLM, TXNRD1, NQO1, PIR and AKR1C1 were also induced in the bronchial airways of smokers compared to non-smokers, whereas MMP10 is repressed in both (Table 3). Pathways and functional categories consistently altered in both groups are given in Supplementary Tables 4 and 5.
Table 3.
Genes that are differentially expressed in both the airways of human smokers and MSK-Leuk1 cells exposed to TS for 0.5–24 hr with associated P-values.
| Symbol | ID | Human | 0.5 hr | 3hr | 6hr | 12 hr | 24 hr | Description |
|---|---|---|---|---|---|---|---|---|
| Airway | p | p | p | p | p | |||
| CYP1A1 | 205749_at | ↑4.4E-04 | ↑3.3E-06 | ↑2.1E-08 | ↑1.1E-07 | ↑1.5E-06 | ↑1.0E-05 | cytochrome P450, family 1, subfam. A, polypep. 1 |
| CYP1B1 | 202436_s_at | ↑1.7E-07 | ↑4.3E-06 | ↑4.5E-05 | ↑5.5E-08 | ↑1.5E-06 | ↑6.6E-04 | cytochrome P450, family 1, subfam. B, polypep. 1 |
| CYP1B1 | 202437_s_at | ↑8.5E-08 | ↑4.2E-06 | ↑4.0E-06 | ↑1.7E-06 | ↑5.5E-06 | ↑2.1E-08 | cytochrome P450, family 1, subfam. B, polypep. 1 |
| GCLM | 203925_at | ↑1.8E-05 | ↑1.5E-03 | ↑1.3E-03 | ↑3.4E-04 | ↑1.8E-04 | ↑3.5E-03 | glutamate-cysteine ligase, modifier subunit |
| TXNRD1 | 201266_at | ↑6.0E-07 | ↑1.2E-02 | ↑2.7E-04 | ↑4.9E-05 | ↑8.8E-05 | ↑1.6E-03 | thioredoxin reductase 1 |
| ALDH1A3 | 203180_at | ↑1.5E-04 | ↑1.9E-04 | ↑9.8E-09 | ↑2.8E-06 | ↑1.3E-03 | ↑2.8E-04 | aldehyde dehydrogenase 1 family, member A3 |
| NQO1 | 201468_s_at | ↑2.8E-13 | ↑4.7E-05 | ↑2.3E-05 | ↑1.3E-06 | ↑6.6E-04 | ↑1.8E-04 | NAD(P)H dehydrogenase, quinone 1 |
| NQO1 | 201467_s_at | ↑4.7E-12 | 3.2E-02 | >0.05 | ↑9.0E-05 | ↑4.0E-03 | ↑4.8E-03 | NAD(P)H dehydrogenase, quinone 1 |
| PIR | 207469_s_at | ↑1.3E-12 | >0.05 | >0.05 | ↑2.8E-02 | ↑3.1E-02 | ↑3.2E-02 | Pirin |
| AKR1C1 | 204151_x_at | ↑1.4E-10 | ↓2.8E-02 | >0.05 | ↑3.8E-03 | ↑1.5E-02 | ↑6.0E-04 | aldo-keto reductase family 1, member C1 |
| NQO1 | 210519_s_at | ↑3.1E-14 | ↓4.7E-02 | ↑1.3E-02 | ↑2.0E-05 | ↑2.1E-03 | ↑3.7E-04 | NAD(P)H dehydrogenase, quinone 1 |
| AKR1C1 | 216594_x_at | ↑1.2E-09 | >0.05 | >0.05 | ↑2.4E-04 | ↑8.1E-03 | ↑9.3E-03 | aldo-keto reductase family 1, member C1 |
| AKR1C2 | 211653_x_at | ↑4.0E-12 | >0.05 | >0.05 | ↑9.6E-03 | ↑2.3E-02 | ↑1.6E-04 | aldo-keto reductase family 1, member C2 |
| AKR1C2 | 209699_x_at | ↑2.5E-14 | ↑3.8E-02 | >0.05 | ↑2.5E-03 | >0.05 | ↑4.4E-04 | aldo-keto reductase family 1, member C2 |
| MMP10 | 205680_at | ↓5.6E-04 | >0.05 | >0.05 | ↓2.2E-02 | >0.05 | >0.05 | matrix metalloproteinase 10 (stromelysin 2) |
We further explored the similarity between the gene expression datasets from airway epithelia and the MSK-Leuk1 cells, by comparing for relative enrichment using the GSEA method (27) as described in the Methods section. At every time point, the TS-treated MSK-Leuk1 gene set was found to be significantly enriched in genes upregulated in airway epithelial cells (FDR <0.001); a significant negative correlation was observed in this analysis with the downregulated genes (FDR = 0.019, 0.001, 0.037, 0.004, 0.01 for 0.5 hr, 3 hr, 6 hr, 12 hr and 24 hr, respectively). In the reverse analysis, the airway data set was found to be significantly enriched in genes induced in MSK-Leuk1 cells at every time point (FDR = 0.043, 0.009, 0.011, 0.009, 0.048 for 0.5 hr, 3hr, 6 hr, 12 hr and 24 hr, respectively). Thus, the results from this Gene Set Enrichment analysis identify significant correlations between the sets of data from the two experiments, and further support the use of the MSK-Leuk1 cell line as a model for detailed mechanistic studies on tobacco smoke effects.
The leading-edge subsets of the significant gene sets in airway epithelia include 50 upregulated and 16 downregulated genes. Of these, ALDH1A3, CYP1A1, CYP1B1, GCLM, GPX2, NQO1, PIR, SERPINB13, SLC7A11, TXNDR1 and UGT1A10 /// UGT1A8 are members of a consistently induced subset (in at least 4 time points). At the same time, KAL1, MMP10, NFKBIA, TMEM45A are members of a consistently repressed subset (in at least 4 time points) in both sets of experiments. Interestingly, the highest number of leading edge subset genes in the airway epithelial data are for the 0.5 hr time point, which is interesting because this time point does not have the largest number of differentially expressed genes. The reverse analysis, for the leading edge subsets of the significantly upregulated genes in MSK-Leuk1 data, results in a total of 20 genes enriched in airway epithelial cells. The complete set of results is given in Supplementary Table 6. Details of this analysis and the related html files are available at http://physiology.med.cornell.edu/go/smoke.
Discussion
We characterized here the effects of TS on gene expression and cellular pathways in MSK-Leuk1 cells. As these were shown to relate to xenobiotic metabolism, cell proliferation, apoptosis, and cell movement, the results can potentially bring a new level of understanding to the complex biological effects of tobacco smoke, and in particular to the manner cellular mechanisms are perturbed leading to carcinogenesis. Cancer patients who continue to smoke have a worse prognosis than individuals who quit smoking, but the underlying mechanisms are poorly understood. The specific pathways shown here to be affected, as well as the identified hubs in the networks composed of the differentially expressed genes should provide not only insights into putative mechanisms by which tobacco smoke impacts on carcinogenesis, but also the manner in which it might affect cancer treatment outcome. Of special note in this context is that tobacco smoke induced CYP1A1 and CYP1B1 both in vitro and in vivo. More specifically, increased levels of CYP1A1 and CYP1B1 were found in MSK-Leuk1 cells exposed to TS and in the oral mucosa of humans who smoked cigarettes heavily, results that confirm and amplify a previous report of elevated levels of CYP1B1 in exfoliated buccal mucosal cells from smokers (42). Because both CYP1A1 and CYP1B1 convert a broad array of carcinogens to active metabolites that can form DNA adducts, it becomes important to consider the potential implications of TS-mediated induction of these enzymes. Thus, several classes of carcinogens, e.g. polycyclic aromatic hydrocarbons (PAHs), nitroaromatics, and arylamines are activated to mutagenic derivatives by these enzymes (43, 44). It is possible that TS-mediated induction of CYP1A1 and CYP1B1 will increase the mutagenic effects of these carcinogens. The potential significance of such an effect is underscored by the finding that B[a]P diol epoxide, a mutagen formed by CYP1A1 or CYP1B1, causes adducts along exons of the TP53 gene that correspond to p53 hotspots in human tumors (45). Based on these results, one potential chemopreventive strategy would be to identify agents that suppress tobacco smoke mediated induction of CYP1A1 and CYP1B1.
In addition to being able to activate carcinogens, CYP1A1 and CYP1B1 play a role in the metabolism of several anticancer drugs including docetaxel, tamoxifen and erlotinib (6, 46). Recently, erlotinib, a small molecule inhibitor of EGFR tyrosine kinase, was found to be more effective in the treatment of patients with non-small cell lung cancer who were never smokers compared to smokers (10). Reduced levels of erlotinib were found in the plasma of smokers compared with never smokers suggesting increased metabolic clearance (6). Our finding that levels of CYP1A1 and CYP1B1 are increased in the oral mucosa of smokers raises the distinct possibility that local in addition to systemic clearance of erlotinib will be enhanced in smokers leading to decreased clinical benefit. Collectively, these findings underscore the need for both more careful monitoring of smoking status in clinical trials and increased smoking cessation efforts in cancer patients. Studies are underway or being planned to evaluate the efficacy of erlotinib in the prevention and treatment of head and neck squamous cell carcinoma.
The induction of CYP1A1 and CYP1B1 by TS is consistent with evidence that the AhR plays a central role in regulating these genes. The AhR had been linked to carcinogenesis (47, 48), and the PAHs in tobacco smoke bind to and activate the AhR resulting in the induction of CYP1A1 and CYP1B1 (13, 38, 41). Following ligand-induced activation by PAHs, the AhR releases its chaperoning heat shock protein 90, translocates into the nucleus and dimerizes with AhR nuclear translocator (ARNT) (49). The heterodimer binds to xenobiotic response elements present in the 5’ flanking region of target genes and thereby modulates transcription. In addition to CYP1A1 and CYP1B1, PAHs induce the phase II xenobiotic metabolizing enzyme NQO1 [NAD(P)H: quinone oxidoreductase] and the AhR repressor (AhRR). The AhR and AhRR are believed to constitute a negative feedback loop of xenobiotic signal transduction. The liganded AhR induces AhRR transcription, whereas expressed AhRR, in turn, inhibits the function of AhR (49). Both NQO1 and AhRR were induced in the TS treated MSK-Leuk1 cells although the magnitude of this induction was less than for CYP1A1 and CYP1B1. Clearly, these results suggest that it will be worthwhile to determine if increased levels of NQO1 and AhRR also occur in the buccal mucosa of smokers. Easily accessible buccal mucosa may serve as a surrogate tissue for understanding the effects of tobacco smoke on the biology of difficult to obtain bronchial mucosa. In support of this notion, we found that TS induced changes in the transcriptome of MSK-Leuk1 cells that were mimicked by differences in bronchial mucosa of smokers vs. non-smokers. AhR-driven gene induction observed in the MSK-Leuk1 model matches the in vivo results in airway epithelial cells of smokers (Table 3). In this regard, the fact that a small subset (CYP1A1 and CYP1B1) of highly inducible genes was also overexpressed in the buccal mucosa of smokers is of interest. Future studies of the transcriptome of buccal mucosa of smokers vs. never smokers will be needed to draw more definitive conclusions about how closely the biology of the buccal mucosa reflects changes in the lower respiratory tract.
Our pathway analysis showed that treatment of MSK-Leuk1 cells altered the expression of genes involved in cellular proliferation, raising the possibility that tobacco smoke amplifies its own mutagenicity by stimulating the proliferation of cells, because conversion of tobacco smoke induced DNA adducts to mutations can only occur in proliferating cells (50). Enhanced cell proliferation has been observed in the aerodigestive tracts of active smokers (51). Interestingly, intracellular levels of glutathione, an antioxidant, have been shown to modulate the effects of tobacco smoke condensate on cell proliferation (52). Notably, we find that glutathione metabolism pathway is induced both in MSK-Leuk1 cells and in the airway epithelial cells of smokers.
We and others found that TS-mediated activation of EGFR signaling led to increased cell proliferation (36, 37). In the current study, treatment with TS led to increased levels of both EGFR and its ligands including AREG, TGFA, HB-EGF and EREG, forming an EGFR-centered hub within the interactome maps. Collectively, these findings strengthen the rationale for evaluating whether an inhibitor of EGFR tyrosine kinase can prevent or delay the onset of tobacco smoke-related malignancies of the aerodigestive tract.
Our pathway analysis suggests that TS modulates cell movement, apoptosis, immune function and coagulation. Both inflammation and immune suppression have been suggested to contribute to tobacco smoke induced carcinogenesis (53). Consistently, we identified the differential expression of several interleukins and their receptors, including IL1A, IL1B, IL1RN, IL1R1, IL1R2, IL6, IL7R, IL8, IL11, IL17RC, IL20 and IL24, in addition to TNFA, where IL1B, IL1R1, IL1RN and IL8 are connected within a network. Our pathway-level analysis also links TS exposure to the induction of specific inflammatory pathways (Table 2).
Taken together, our results provide new insights into the potential mechanisms underlying procarcinogenic effects of tobacco smoke and may help to explain the worse outcome of cancer patients who continue to smoke. Furthermore, our results suggest important targets for future studies designed to identify agents that could reduce or eliminate the detrimental effects of TS exposure indicated by the findings presented here.
Supplementary Material
Acknowledgments
We thank Jenny Xiang from the Microarray Core of Weill Cornell Medical College for expert help and Kevin C. Dorff from the Institute for Computational Biomedicine at Cornell University for webpage design. This study was supported in part by resources from the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, NIH T32 CA09685 and the Center for Cancer Prevention Research.
References
- 1.Gritz ER, Dresler C, Sarna L. Smoking, the missing drug interaction in clinical trials: ignoring the obvious. Cancer Epidemiol Biomarkers Prev. 2005;14:2287–2293. doi: 10.1158/1055-9965.EPI-05-0224. [DOI] [PubMed] [Google Scholar]
- 2.Tobacco smoke and involuntary smoking. IARC Monogr Eval Carcinog Risks Hum. 2004;83:1–1438. [PMC free article] [PubMed] [Google Scholar]
- 3.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 4.Mayne ST, Lippman SM. Cigarettes: a smoking gun in cancer chemoprevention. J Natl Cancer Inst. 2005;97:1319–1321. doi: 10.1093/jnci/dji306. [DOI] [PubMed] [Google Scholar]
- 5.The Alpha-Tocopherol, Beta Carotene Cancer Prevention Study Group. The effect of vitamin E beta carotene on the incidence of lung cancer and other cancers in male smokers. N Engl J Med. 1994;330:1029–1035. doi: 10.1056/NEJM199404143301501. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton M, Wolf JL, Rusk J, et al. Effects of smoking on the pharmacokinetics of erlotinib. Clin Cancer Res. 2006;12:2166–2171. doi: 10.1158/1078-0432.CCR-05-2235. [DOI] [PubMed] [Google Scholar]
- 7.Khuri FR, Lee JJ, Lippman SM, et al. Randomized phase III trial of low-dose isotretinoin for prevention of second primary tumors in stage I and II head and neck cancer patients. J Natl Cancer Inst. 2006;98:441–450. doi: 10.1093/jnci/djj091. [DOI] [PubMed] [Google Scholar]
- 8.Fox JL, Rosenzweig KE, Ostroff JS. The effect of smoking status on survival following radiation therapy for non-small cell lung cancer. Lung Cancer. 2004;44:287–293. doi: 10.1016/j.lungcan.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Pantarotto J, Malone S, Dahrouge S, Gallant V, Eapen L. Smoking is associated with worse outcomes in patients with prostate cancer treated by radical radiotherapy. BJU Int. 2007;99:564–569. doi: 10.1111/j.1464-410X.2006.06656.x. [DOI] [PubMed] [Google Scholar]
- 10.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated nonsmall-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 11.Browman GP, Wong G, Hodson I, et al. Influence of cigarette smoking on the efficacy of radiation therapy in head and neck cancer. N Engl J Med. 1993;328:159–163. doi: 10.1056/NEJM199301213280302. [DOI] [PubMed] [Google Scholar]
- 12.Guengerich FP, Shimada T. Activation of procarcinogens by human cytochrome P450 enzymes. Mutat Res. 1998;400:201–213. doi: 10.1016/s0027-5107(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 13.Murray GI, Melvin WT, Greenlee WF, Burke MD. Regulation, function, and tissue-specific expression of cytochrome P450 CYP1B1. Annu Rev Pharmacol Toxicol. 2001;41:297–316. doi: 10.1146/annurev.pharmtox.41.1.297. [DOI] [PubMed] [Google Scholar]
- 14.Ko Y, Abel J, Harth V, et al. Association of CYP1B1 codon 432 mutant allele in head and neck squamous cell cancer is reflected by somatic mutations of p53 in tumor tissue. Cancer Res. 2001;61:4398–4404. [PubMed] [Google Scholar]
- 15.Spira A, Beane J, Shah V, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci U S A. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sacks PG. Cell, tissue and organ culture as in vitro models to study the biology of squamous cell carcinomas of the head and neck. Cancer Metastasis Rev. 1996;15:27–51. doi: 10.1007/BF00049486. [DOI] [PubMed] [Google Scholar]
- 17.Moraitis D, Du B, De Lorenzo MS, et al. Levels of cyclooxygenase-2 are increased in the oral mucosa of smokers: evidence for the role of epidermal growth factor receptor and its ligands. Cancer Res. 2005;65:664–670. [PubMed] [Google Scholar]
- 18.Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005;6:R16. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z, Irizarry RA, Gentleman RFM, Spencer F. A model based background adjustment for oligonucleotide expression arrays. J Amer Stat Assoc. 2003;99:909–917. [Google Scholar]
- 21.Shedden K, Chen W, Kuick R, et al. Comparison of seven methods for producing Affymetrix expression scores based on False Discovery Rates in disease profiling data. BMC Bioinformatics. 2005;6:26. doi: 10.1186/1471-2105-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cope LM, Irizarry RA, Jaffee HA, Wu Z, Speed TP. A benchmark for Affymetrix GeneChip expression measures. Bioinformatics. 2004;20:323–331. doi: 10.1093/bioinformatics/btg410. [DOI] [PubMed] [Google Scholar]
- 23.Millenaar FF, Okyere J, May ST, et al. How to decide? Different methods of calculating gene expression from short oligonucleotide array data will give different results. BMC Bioinformatics. 2006;7:137. doi: 10.1186/1471-2105-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 26.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhodes DR, Tomlins SA, Varambally S, et al. Probabilistic model of the human protein-protein interaction network. Nat Biotechnol. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- 29.http://www.ingenuity.com [cited 2007 Dec 15].
- 30.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomfohr J, Lu J, Kepler TB. Pathway level analysis of gene expression using singular value decomposition. BMC Bioinformatics. 2005;6:225. doi: 10.1186/1471-2105-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21:1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- 33.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma P, Castillo-Davis CI, Zhong W, Liu JS. A data-driven clustering method for time course gene expression data. Nucleic Acids Res. 2006;34:1261–1269. doi: 10.1093/nar/gkl013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan MP, Broach JR, Floudas CA. A novel clustering approach and prediction of optimal number of clusteris: I. Global Optimum Search with Enhanced Positioning. J Global Optimization. 2007;39:323–346. doi: 10.1142/s0219720007002941. [DOI] [PubMed] [Google Scholar]
- 36.Du B, Altorki NK, Kopelovich L, Subbaramaiah K, Dannenberg AJ. Tobacco smoke stimulates the transcription of amphiregulin in human oral epithelial cells: evidence of a cyclic AMP-responsive element binding protein-dependent mechanism. Cancer Res. 2005;65:5982–5988. doi: 10.1158/0008-5472.CAN-05-0628. [DOI] [PubMed] [Google Scholar]
- 37.Lemjabbar H, Li D, Gallup M, et al. Tobacco smoke-induced lung cell proliferation mediated by tumor necrosis factor alpha-converting enzyme and amphiregulin. J Biol Chem. 2003;278:26202–26207. doi: 10.1074/jbc.M207018200. [DOI] [PubMed] [Google Scholar]
- 38.Port JL, Yamaguchi K, Du B, et al. Tobacco smoke induces CYP1B1 in the aerodigestive tract. Carcinogenesis. 2004;25:2275–2281. doi: 10.1093/carcin/bgh243. [DOI] [PubMed] [Google Scholar]
- 39.Du B, Leung H, Khan KM, et al. Tobacco smoke induces urokinase-type plasminogen activator and cell invasiveness: evidence for an epidermal growth factor receptor dependent mechanism. Cancer Res. 2007;67:8966–8972. doi: 10.1158/0008-5472.CAN-07-1388. [DOI] [PubMed] [Google Scholar]
- 40.Peri S, Navarro JD, Amanchy R, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brauze D, Widerak M, Cwykiel J, Szyfter K, Baer-Dubowska W. The effect of aryl hydrocarbon receptor ligands on the expression of AhR, AhRR, ARNT, Hif1alpha, CYP1A1 and NQO1 genes in rat liver. Toxicol Lett. 2006;167:212–220. doi: 10.1016/j.toxlet.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 42.Spivack SD, Hurteau GJ, Jain R, et al. Gene-environment interaction signatures by quantitative mRNA profiling in exfoliated buccal mucosal cells. Cancer Res. 2004;64:6805–6813. doi: 10.1158/0008-5472.CAN-04-1771. [DOI] [PubMed] [Google Scholar]
- 43.Josephy PD, Batty SM, Boverhof DR. Recombinant human P450 forms 1A1, 1A2, and 1B1 catalyze the bioactivation of heterocyclic amine mutagens in Escherichia coli lacZ strains. Environ Mol Mutagen. 2001;38:12–18. doi: 10.1002/em.1045. [DOI] [PubMed] [Google Scholar]
- 44.Shimada T, Hayes CL, Yamazaki H, et al. Activation of chemically diverse procarcinogens by human cytochrome P-450 1B1. Cancer Res. 1996;56:2979–2984. [PubMed] [Google Scholar]
- 45.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 46.McFadyen MC, McLeod HL, Jackson FC, et al. Cytochrome P450 CYP1B1 protein expression: a novel mechanism of anticancer drug resistance. Biochem Pharmacol. 2001;62:207–212. doi: 10.1016/s0006-2952(01)00643-8. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu Y, Nakatsuru Y, Ichinose M, et al. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2000;97:779–782. doi: 10.1073/pnas.97.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersson P, McGuire J, Rubio C, et al. A constitutively active dioxin/aryl hydrocarbon receptor induces stomach tumors. Proc Natl Acad Sci U S A. 2002;99:9990–9995. doi: 10.1073/pnas.152706299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mimura J, Fujii-Kuriyama Y. Functional role of AhR in the expression of toxic effects by TCDD. Biochim Biophys Acta. 2003;1619:263–268. doi: 10.1016/s0304-4165(02)00485-3. [DOI] [PubMed] [Google Scholar]
- 50.Cohen SM, Ellwein LB. Cell proliferation in carcinogenesis. Science. 1990;249:1007–1011. doi: 10.1126/science.2204108. [DOI] [PubMed] [Google Scholar]
- 51.van Oijen MG, Gilsing MM, Rijksen G, Hordijk GJ, Slootweg PJ. Increased number of proliferating cells in oral epithelium from smokers and ex-smokers. Oral Oncol. 1998;34:297–303. doi: 10.1016/s1368-8375(98)00007-4. [DOI] [PubMed] [Google Scholar]
- 52.Luppi F, Aarbiou J, van Wetering S, et al. Effects of cigarette smoke condensate on proliferation and wound closure of bronchial epithelial cells in vitro: role of glutathione. Respir Res. 2005;6:140. doi: 10.1186/1465-9921-6-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ballaz S, Mulshine JL. The potential contributions of chronic inflammation to lung carcinogenesis. Clin Lung Cancer. 2003;5:46–62. doi: 10.3816/CLC.2003.n.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.