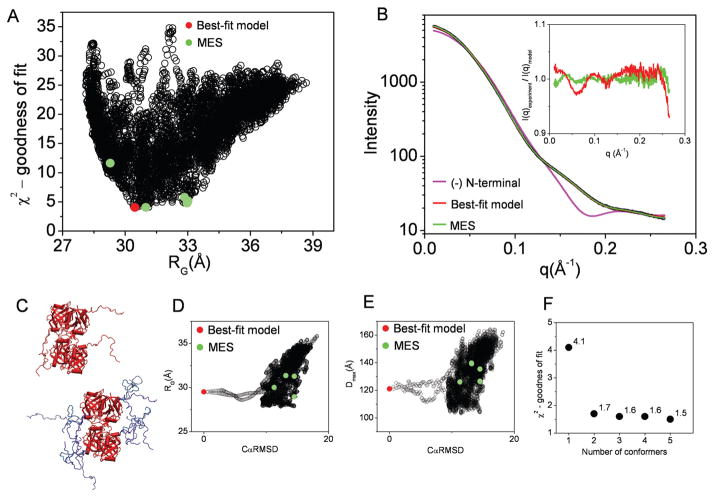
Figure 6.
BILBOMD analysis of Flavin reductase domain protein (FRDP). A. Graph represents the comparison of discrepancy (χ2) values for 7000 FRDP models with their RG values. The value for best-fit model is indicated by red dot. The green dots indicate the five MES conformers. B. Experimental SAXS curve (black). The theoretical scattering profile for FRDP atomic structure missing flexible N-terminal extension (pink line). Red and green lines represent the theoretical scattering for the best-fit and MES ensemble, respectively. Inset – calculated residuals (I(q)experiment/I(q)model) for MES ensemble with 5 conformers and single best-fit model. C. The best-fit model for FRDP (left panel). The five MES-selected models (right panel). D., E. Graph represents the comparison of RG and Dmax values for all 7000 models with their CαRMSD values referenced to the best-fit model. The values for the best-fit model and MES-selected models are indicated by red and green dots, respectively. F. χ2 values for MES fit in dependence to the number of selected conformers.