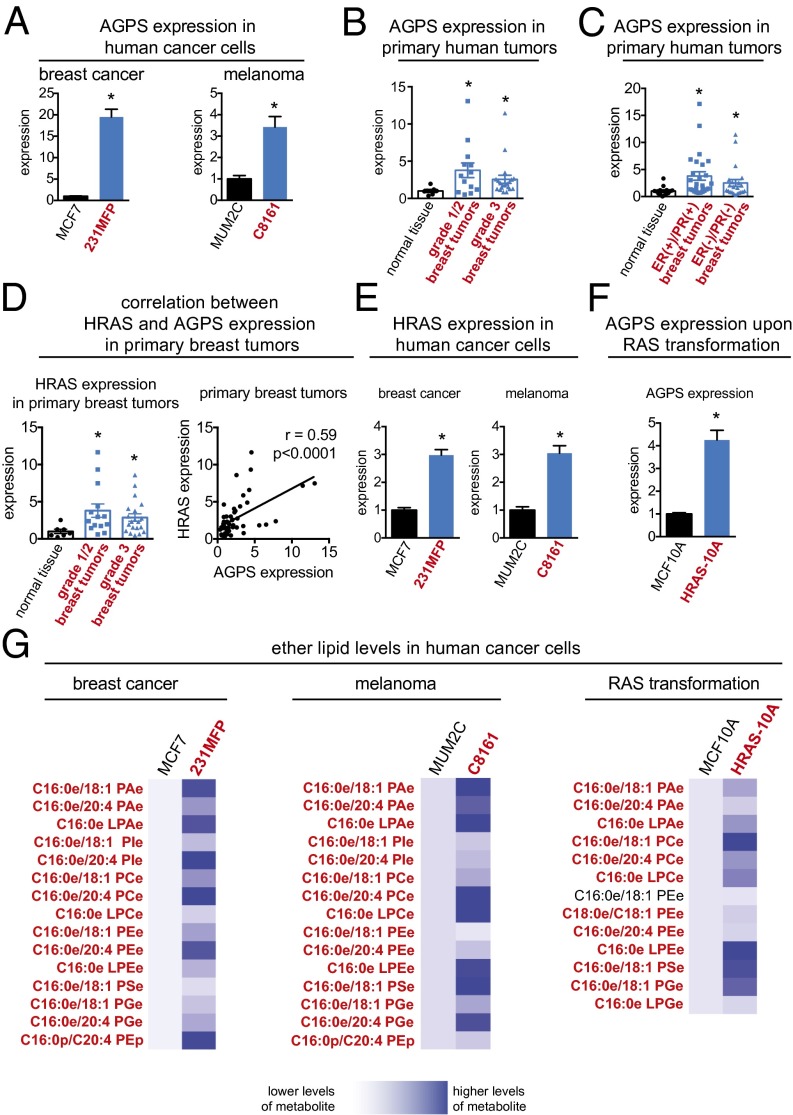
Fig. 1.
AGPS is highly expressed in aggressive cancer cells, primary human tumors, and RAS-transformed cells. (A) AGPS gene expression is heightened across aggressive breast (231MFP) and melanoma (C8161) cancer cells (in red) compared with less aggressive breast (MCF7) and melanoma (MUM2C) cancer cells (in black), as measured by qPCR. (B) AGPS gene expression is significantly higher in Nottingham grade 1 (low-grade) and grade 2 (intermediate-grade), as well as grade 3 (high-grade) primary human breast tumors compared with normal breast tissue as measured by qPCR. (C) AGPS gene expression is also significantly higher in ER(+)/PR(+) and ER(−)/PR(−) human breast tumors compared with normal breast tissue. (D) HRAS expression is higher in grade 1/2 and 3 primary human breast tumors compared with normal breast tissue. HRAS expression from matching normal tissue and breast tumors is significantly correlated with AGPS expression with a Pearson correlation coefficient of r = 0.59. (E) HRAS expression is higher in aggressive breast and melanoma cancer cells compared with less aggressive cells. (F) HRAS overexpression in MCF10A nontransformed mammary epithelial cells increases AGPS expression. (G) Aggressive breast and melanoma cancer cells and HRAS-transformed MCF10A cells possess significantly higher levels of multiple species of ether lipids compared with less aggressive or empty vector-infected MCF10A control cells, respectively. Data in A–F are presented as mean ± SEM; n = 4 per group for A, E, and F, n = 7–26 per group for B–D, and n = 4–5 per group for G. Significance in A–F is presented as *P < 0.05 compared with less-aggressive cancer cells (A and E), normal breast tissue (B–D), or empty vector-infected MCF10A cells (F). Significance in G is presented as P < 0.05 for lipid designations that are bolded in red comparing 231MFP to MCF7, C8161 to MUM2C, or HRAS-10A to MCF10A groups.