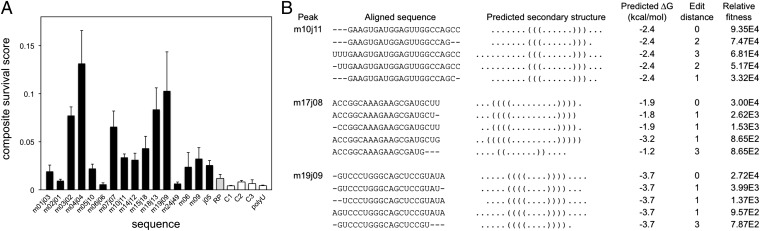
Fig. 4.
Fitness and secondary structures for individual RNA sequences. (A) Several peak sequences (black bars), the initial pool (gray bar), and negative control sequences (white bars) were assayed individually for survival through the negative selection (agarose resin) and positive selection (GTP agarose resin) steps as well as relative enrichment during PCR. Relative fitnesses through each step and the expected ligation bias were combined into a composite survival score (Materials and Methods and SI Appendix, Table S5); error bars represent the SD. (B) Predicted secondary structures for the three peaks with lowest minimum free energy. The five sequences with highest fitness from each peak are shown with aligned secondary structures in bracket notation. Edit distance from the peak center and the relative fitness of each sequence are also shown.