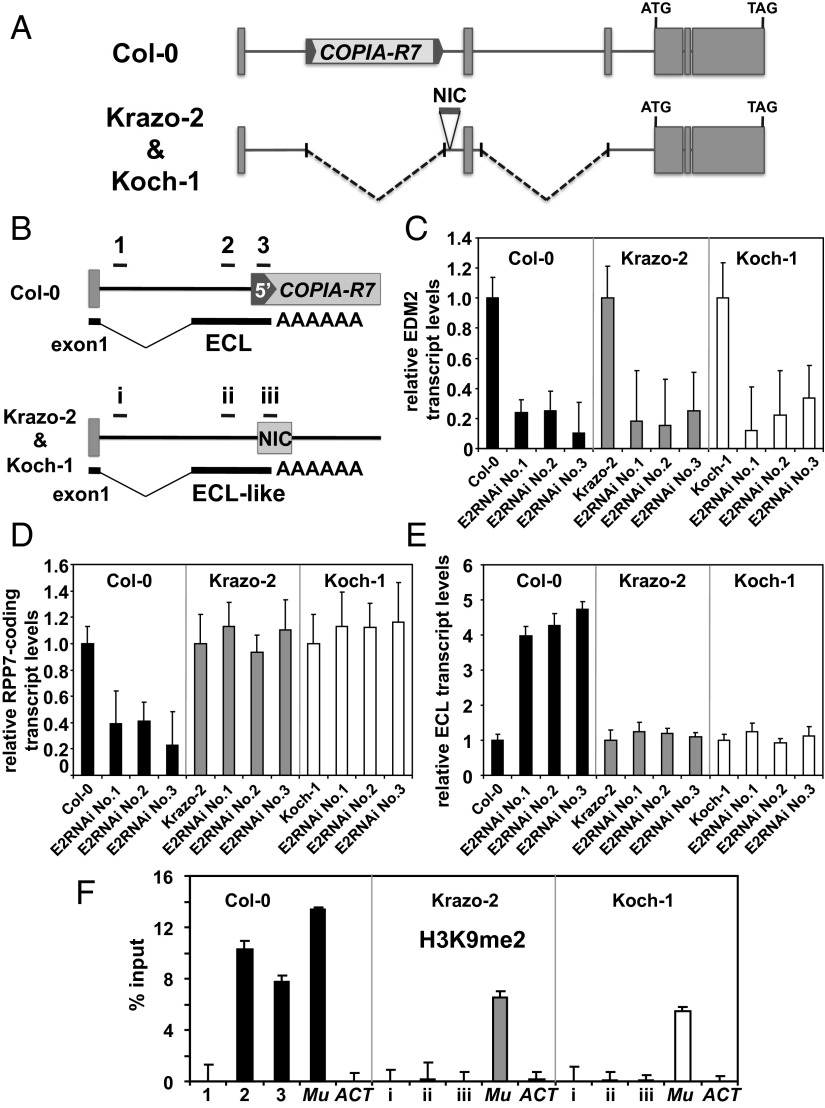
Fig. 5.
Effects of EDM2 on RPP7 expression in Arabidopsis natural accessions. (A) Schematic representation of the RPP7 locus in Arabidopsis Col-0, Krazo-2, and Koch-1 accessions. RPP7 exons are shown as gray-filled boxes. The genomic DNA sequence block NIC in Krazo-2/Koch-1 is represented as gray-filled horizontal bars. (B) Schematic representation of the upstream region of RPP7 locus with the RNA transcript isoform ECL in Col-0 and Krazo-2/Koch-1. Horizontal bars (1–3 and i–iii) denote regions analyzed by ChIP for H3K9me2 in F. (C) Transcript levels of EDM2 determined by qRT-PCR in Col-0, Krazo-2, Koch-1, and EDM2 silencing lines in the background of each of these three Arabidopsis accessions. Error bars represent SD for three independent experiments. (D) Levels of RPP7-coding transcripts determined by qRT-PCR in Col-0, Krazo-2, Koch-1, and the respective EDM2 silencing lines. Error bars represent SD for three independent experiments. (E) Levels of ECL transcripts determined by qRT-PCR in Col-0, Krazo-2, Koch-1, and the respective EDM2 silencing lines. Error bars represent SD for three independent experiments. (F) ChIP-qPCR to measure H3K9me2 levels at RPP7 in Col-0, Krazo-2, and Koch-1. The y axis represents H3K9me2 levels normalized to input DNA. The ECL regions represented in B were tested. The DNA transposon Mu1 (Mu) and Actin8 (ACT) served as positive and negative controls, respectively. Error bars represent SEM for two biological replicates with three technical replicates each.