Abstract
Ferric enterobactin (FeEnt) acquisition plays a critical role in the pathophysiology of Campylobacter, the leading bacterial cause of human gastroenteritis in industrialized countries. In Campylobacter, the surface-exposed receptor, CfrA or CfrB, functions as a “gatekeeper” for initial binding of FeEnt. Subsequent transport across the outer membrane is energized by TonB-ExbB-ExbD energy transduction systems. Although there are up to three TonB-ExbB-ExbD systems in Campylobacter, the cognate components of TonB-ExbB-ExbD for FeEnt acquisition are still largely unknown. In this study, we addressed this issue using complementary molecular approaches including: comparative genomic analysis, random transposon mutagenesis, and site-directed mutagenesis in two representative C. jejuni strains, NCTC 11168 and 81-176. We demonstrated that CfrB could interact with either TonB2 or TonB3 for efficient Ent-mediated iron acquisition. However, TonB3 is a dominant player in CfrA-dependent pathway. The ExbB2 and ExbD2 components were essential for both CfrA- and CfrB-dependent FeEnt acquisition. Sequences analysis identified potential TonB boxes in CfrA and CfrB, and the corresponding binding sites in TonB. In conclusion, these findings reveal identities of specific TonB-ExbB-ExbD energy transduction components required for FeEnt acquisition, and provide insights into the complex molecular interactions of FeEnt acquisition systems in Campylobacter.
Keywords: Iron uptake, siderophore, molecular mechanism
Introduction
Higher organisms have evolved complicated mechanisms for sequestrating free iron to well below those required for the growth of Gram-negative bacteria (Braun, et al., 1998). To counteract iron-limitation for successful in vivo colonization, Gram-negative bacteria have evolved complex and aggressive genetic systems for iron uptake (Braun, et al., 1998, Andrews, et al., 2003, Wandersman & Delepelaire, 2004, Miethke & Marahiel, 2007). For most of the iron uptake systems, iron-regulated outer membrane proteins function as the first line to recognize and bind specific iron complex. Following binding, specific iron complex must be transported through the receptor channel using TonB-ExbB-ExbD energy transduction system, in which TonB is an inner membrane-anchored, periplasm-spanning protein while ExbB and ExbD proteins are embedded in inner membrane (Raymond, et al., 2003, Miethke & Marahiel, 2007). The TonB-ExbB-ExbD system transduces the proton motive force energy to the receptor to allow translocation of specific iron source. Therefore, the three-component TonB complex plays an essential role in bacterial iron acquisition.
As the most efficient and common iron scavenging mechanism in Gram-negative bacteria, siderophore-mediated iron acquisition has drawn extensive attentions (Miethke & Marahiel, 2007). In particular, enterobactin (Ent)-mediated iron acquisition has been widely investigated because of the extremely high affinity of Ent to ferric iron (Raymond, et al., 2003) and physiological relevance of Ent utilization for bacterial pathogens (Palyada, et al., 2004, Xu, et al., 2010, Pi, et al., 2012). Recently, we identified and characterized two ferric enterobactin (FeEnt) receptors, CfrA and CfrB in Campylobacter, the leading bacterial cause of human gastroenteritis in the United States and industrialized countries (Zeng, et al., 2009, Xu, et al., 2010). Interestingly, analysis of published genome of C. jejuni NCTC 11168 (Parkhill, et al., 2000) reveals three sets of TonB-ExbB-ExbD systems, which are all subjected iron regulation (Palyada, et al., 2004). Briefly, the tonB1 (Cj0181) and tonB2 (Cj1630) are organized into the same operon with their corresponding exbB/exbD genes (Cj0179/Cj0180 and Cj1628/Cj1629, respectively). The tonB3 (Cj0753c) is an ‘orphan’ gene with no adjacent exbB/exbD genes. There exists a third pair of exbB/exbD (Cj0109/Cj0110) in NCTC 11168 genome, which was designated as exbB3/exbD3 despite their distant location with the tonB3. In this study, we performed genomics and molecular studies to identify the cognate TonB, ExbB, and ExbD components required for CfrA- and CfrB-dependent FeEnt acquisition. The findings from this study provide insights into the molecular interactions and evolution of FeEnt acquisition systems in Campylobacter.
Materials and Methods
Bacterial strains, plasmids, and culture condition
The major bacterial strains and plasmids used in this study are listed in Table 1. In general, C. jejuni strains were cultivated in Müller-Hinton (MH) broth or on agar at 42°C under microaerophilic conditions (85% N2, 10% CO2, 5% O2). To achieve iron-restricted conditions, 20 μM of deferoxamine mesylate (DFO) was added into media. E. coli strains were grown routinely in Luria-Bertani (LB) broth with shaking (250 rpm) or on agar at 37 °C overnight. When needed, culture media were supplemented with ampicillin (100 μg/ml), kanamycin (30 μg/ml), chloramphenicol (Cm) (6 μg/ml for Campylobacter and 20 μg/ml for E. coli), erythromycin (5 μg/ml for Campylobacter and 200 μg/ml for E. coli) or tetracycline (5 μg/ml for Campylobacter and 12.5 μg/ml for E. coli).
Table 1.
Key bacterial plasmids and strains used in this study
| Plasmids or strains | Description | Source or Reference |
|---|---|---|
| Plasmids | ||
| pGEMT-Easy | PCR cloning vector, Ampr | Promega |
| pCee | T-easy vector derivative containing Cj1376 and partial Cj1377c | (Zeng, et al., 2013) |
| pCee+Cm | pCee derivative with cat cassette inserted at PacI site between cee and cj1377c | This study |
| pRR | T-easy vector derivative containing C.jejuni NCTC 11168 ribosomal fragment | This study |
| pRR-CfrB | pRR derivative in which cfrB gene was inserted inside ribosomal region | This study |
| pRRE-CfrB | The erythromycin resistant cassette was attached at the end of cfrB in pRR-CfrB | This study |
| pRY107 | E. coli-Campylobacter shuttle vector, kanamycin resistant (Kmr) | (Yao, et al., 1993) |
| pTonB3 | pRY107 derivative containing the tonB3 and its promoter. | This study |
| pCfrA | pRY107 derivative containing cfrA plus its promoter region | (Zeng, et al., 2009) |
| pTonB3-CfrA | pCfrA derivative containing both intact tonB3 and cfrA genes | This study |
| pTonB3-CfrA-T | T-easy vector derivative containing the tonB3-cfrA locus | This study |
| pTonB3(tetO) | pTonB3-CfrA-T derivative with tetO cassette inserted in tonB3 | This study |
| Strains | ||
| C.jejuni | ||
| 81-176 | Human isolate | (Black, et al., 1988) |
| JL727 | 81-176 derivative for which 81-176 was chromosomally complemented with cee gene | This study |
| JL680 | 81-176 complemented with plasmid pTonB3-CfrA | This study |
| JL534 | 81-176 complemented with plasmid pCfrA | This study |
| 11168 | NCTC 11168, human isolate | (Parkhill, et al., 2000) |
| JL709 | 11168 derivative with cat cassette inserted between cee and cj1377c | This study |
| JL832 | 11168 derivative with tonB3 inactivated by insertion of tetO cassette | This study |
| JL324 | 11168 derivative, cfrA::cat | (Zeng, et al., 2009) |
| JL612 | JL324 derivative with chromosomal complementation of cfrB gene. Thus, this strain has genetically repaired CfrB-dependent pathway for FeEnt acquisition |
This study |
| JL845 | JL612 derivative, tonB2− cfrA− cfrB+ | This study |
| JL868 | JL845 derivative, tonB2− tonB3− cfrA− cfrB+ | This study |
| JL869 | JL845 derivative, tonB3− cfrA− cfrB+ | This study |
| E. coli | ||
| DH5α | F- Φ80lacZΔM15 Δ(lacZYA-argF)U169 recAl endAl hsdR17 (rk−, mk+) phoA supE44 thi-1 gyrA96 relAl λ- |
Invitrogen |
| JL48 | DH5α/pRK2013, help strain for conjugation. | (Zeng, et al., 2009, Xu, et al., 2010) |
Construction of tonB3 mutant
The tonB3 gene was inactivated by allelic exchange using suicide plasmid as described previously (Hofreuter, et al., 2006, Zeng, et al., 2009). Briefly, the 2,388 bp fragment covering tonB3 and its adjacent cfrA was amplified from NCTC 11168 using primers TonB3F (Table 2) and CfrAR2 (Zeng, et al., 2009), and the PCR product was then cloned into pGEMT-Easy (Promega). The resulting plasmid (pTonB3-CfrA-T) was digested with BsrGI and end repaired with T4 DNA polymerase. The tetracycline resistant gene (tetO) was PCR amplified from genomic DNA of C. jejuni 81-176 using Pfu Turbo DNA polymerase (Stratagene) with the primers described in a previous publication (Jeon, et al., 2011); the tetO PCR fragment was then ligated to the BsrGI-treated pTonB3-CfrA-T, creating the suicide vector pTonB3(tetO). Sequence analysis of the construct indicated that the Tet resistance cassette was inserted into tonB3 with the same transcriptional direction. This suicide vector was transferred into NCTC 11168 by natural transformation (Wang & Taylor, 1990). The isogenic tonB3 mutant, named JL832 (Table 1), was selected on MH agar plate containing 5 μg/ml of Tet. The inactivation of the tonB3 in JL832 was confirmed by PCR (data not shown).
Table 2.
Major primers used in this study
| Primer | DNA Sequence (5′-3′)a | Product size (bp) |
Target gene/ operon |
|---|---|---|---|
| Cj1376_F | TTTATCGCTATGGGCTTTGC | 1,917 | C. jejuni cee locus |
| Cj1376_R | TTGCAAAATGTTTTAAAAGAGCA | ||
| Cm_PacI_F | CCCTTAATTAATGCTCGGCGGTGTTCCTTT | 801 | Chloramphenicol resistance cassette |
| Cm_PacI_R | CCCTTAATTAAGCGCCCTTTAGTTCCTAAAG | ||
| TonB3F | TGGCAACACTTTACATAG | 1,343 | tonB3 with promoter |
| TonB3R | CATTGATAGTAGCAGGAG | ||
| rrsF | CTGGAACTCAACTGACGCTAAG | 1,900 | Ribosomal DNA |
| rrlR | CTCTTGCACATTGCAGTCCTAC | ||
| CfrB_XbaI_F | GCTCTAGATGGAGCCTATCAAGAGGCTTAG | 2,409 | cfrB with promoter |
| CfrB_MfeI_R | GCGCAATTGCCAAGTGCAAAGCCTACCAT | ||
| Erm_MfeI_F | GCGCAATTGAGCTTTGGCTAACACACACG | 1,140 | Erythromycin resistance cassette |
| Erm_MfeI_R | GCGCAATTGAATAGGTACACGAAAAACAAGT TAAGG |
Restriction sites are underlined in the primer sequence.
Random transposon mutagenesis
we have successfully used C. jejuni 81-176 as a host strain for in vivo random transposon mutagenesis (Lin, et al., 2009, Hoang, et al., 2011, Hoang, et al., 2012). C. jejuni 81-176 cannot utilize FeEnt unless complemented with a periplasmic trilactone esterase Cee (Zeng, et al., 2013). Therefore, we first constructed an 81-176 derivative (JL727, Table 1) that has Cee complementation in chromosome. Briefly, the Cm resistance gene was amplified from plasmid pUOA18 (Wang & Taylor, 1990) with primers Cm_PacI_F and Cm_PacI_R (Table 2) using PfuUltra® High-Fidelity DNA polymerase (Stratagene). The 0.8-kb PCR product containing a Cm resistance gene was ligated to PacI-digested pCee to obtain vector pCee+Cm in which the Cm resistance gene is inserted between cee and its downstream gene Cj1377c. Then, the pCee+Cm vector was introduced into NCTC 11168 using natural transformation; the resulting Cm resistant mutant (named JL709, Table 1) has Cm resistance gene between cee and Cj1377c in the chromosome of NCTC 11168. The genomic DNA from JL709 was further used for natural transformation with C. jejuni 81-176 as a host strain, creating strain JL727 (Table 1) in which the cee gene together with the Cm resistance cassette was inserted in the chromosome of 81-176. Insertion of the cee and the Cm resistance gene was confirmed by PCR; such chromosomal complementation successfully restored 81-176’s ability to utilize FeEnt as a sole iron source for growth (data not shown).
The JL727 strain was then subjected to the in vivo random transposon mutagenesis; the procedure and screening strategy are detailed in previous publications (Lin, et al., 2009, Hoang, et al., 2011). The mutants with growth defects in MH broth containing kanamycin (50 μg/ml), DFO (20 μM) and Ent (5 μM) were identified. To determine transposon insertion site, genomic DNA extracted from FeEnt-deficient mutants were subjected to direct sequencing as described previously (Lin, et al., 2009, Hoang, et al., 2011).
Chromosomal complementation of CfrB in NCTC 11168
The chromosomal complementation of CfrB in NCTC 11168 was performed using the similar strategy as described previously (Karlyshev & Wren, 2005), in which the target gene is inserted in ribosomal loci. Briefly, the ribosomal region was amplified from NCTC 11168 with primer rrsF and rrlR (Table 2) using GoTaq PCR master mix (Promega). The PCR product was ligated into pGEMT-Easy (Promega), resulting in plasmid pRR. The complete cfrB gene together with its promoter was amplified from C. jejuni 81-176 with primer pairs of CfrB_XbaI_F and CfrB_XbaI_F (Table 2) using PfuUltra® High-Fidelity DNA polymerase (Stratagene). Both pRR and the cfrB fragment were digested with XbaI and MfeI and ligated together, generating pRR-CfrB in which the cfrB gene was inserted inside of the ribosomal region. The erythromycin resistant cassette, erm, was amplified from genomic DNA of the cfrA::Erm mutant of NCTC 11168 (Kindly provided by Dr. Richard D. Haigh, University of Leicester) with primer pairs of Erm_MfeI_F and Erm_MfeI_R using PfuUltra® High-Fidelity DNA polymerase. The erm PCR product was digested with MfeI and was ligated to the pRR-CfrB that was digested with the same restriction enzyme, creating vector pRRE-CfrB in which the erm gene is immediately downstream of cfrB gene with the same transcriptional direction (Table 1). Approximately 10 μg of the pRRE-CfrB was introduced into JL324, an isogenic cfrA mutant of NCTC 11168 (Table 1), by natural transformation. One erythromycin resistant mutant was selected on MH plates containing 5 μg/ml of erythromycin. This mutant, named JL612, has a functional cfrB gene inserted in ribosomal region of chromosome, which was confirmed by PCR (data not shown).
The tonB2 mutation from a TonB2 mutant (1-3D2, Table 4) was introduced into JL612 by natural transformation, creating mutant JL845 (Table 1). Subsequently, the tonB3 gene in JL845 was further inactivated by natural transformation using the genomic DNA from JL832, creating mutant JL868 (Table 1)
Table 4.
EZ::TN™ transposon insertion sites in mutants defective of FeEnt acquisition
| Mutants | Locus (orientationa) |
Tn location (ORF size in bp)b |
Annotation/function |
|---|---|---|---|
| 1-3D2 | tonB2 (−) | 65 (684) | Putative TonB transport protein |
| 4-A1 | tonB2 (+) | 555 (684) | Putative TonB transport protein |
| 4-A5 | exbD2 (+) | 113 (411) | Putative ExbD/TolR family transport protein |
| 4-A11 | exbB2 (+) | 391 (438) | Putative ExbB/TolQ family transport protein |
| 4-D12 | tonB2 (+) | 71 (684) | Putative TonB transport protein |
| 5-C3 | cee (+) | 88 (810) | Periplasmic enterobactin esterase |
| 5-D3 | cee (+) | 459 (810) | Periplasmic enterobactin esterase |
| 5-D4 | cee (+) | 330 (810) | Periplasmic enterobactin esterase |
the orientation of transposon relative to that of disrupted locus. +, same orientation; −, opposite orientation.
The number indicates the nucleotide before which the transposon (Tn) is inserted.
ORF, open reading frame.
Complementation in trans
The ‘orphan’ tonB3 gene was amplified from NCTC 11168 with the primer pairs of TonB3F and TonB3R (Table 2) using PfuUltra DNA polymerase, and then ligated into the SmaI-digested shuttle vector pRY107, generating pTonB3 (Table 1). The pTonB3 was further digested with restriction enzymes PstI and PacI, generating a 1.2-kb fragment containing tonB3; subsequently, this fragment was ligated to the pCfrA (Zeng, et al., 2009) that has been digested with the same restriction enzymes, creating plasmid pTonB3-CfrA. The plasmid pTonB3-CfrA and pCfrA were conjugatively transferred into C. jejuni 81-176 with the help strain JL48, creating complemented constructs JL680 and JL534, respectively (Table 1).
Genomic analysis of TonB-ExbBD
The orthologs shared among finished genomes of C. jejuni NCTC 11168 (NC_002163.1) (Parkhill, et al., 2000), 81-176 (NC_008787.1) (Hofreuter, et al., 2006), and C. coli RM2228 (AAFL01000001-AAFL01000038) (Fouts, et al., 2005) were compared in Xbase (Chaudhuri, et al., 2008). Other Campylobacter genome sequences (finished or draft genomes) were primarily retrieved from CampyDB (http://www.xbase.ac.uk/campydb) or IMG (http://www.hmpdacc-resources.org/cgi-bin/imgm_hmp/main.cgi).
Sequence analysis of CfrA, CfrB, and TonB
The phylogenetical analysis of Campylobacter CfrA, CfrB, and TonB together with their homologs in E. coli were performed in MEGA 5.0 (Kumar, et al., 2008). The phylogenetic tree was constructed using Neighbor-joining methods. To identify the potential TonB box in CfrA and CfrB and the specific binding sites in TonB for TonB box, multiple-sequence alignment of the sequences from Campylobacter as well as E. coli were performed using the ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Results
Comparative genomics analysis of TonB-ExbB-ExbD systems in C. jejuni and C. coli
Analysis of several published C. jejuni genomes has revealed that C. jejuni strains differ in the presence of TonB-ExbB-ExbD systems (Stintzi, et al., 2008, Miller, et al., 2009). In this study, we performed in-depth genomic analysis by taking advantage of the published genome data to date. Table 3 shows the key components of FeEnt acquisition in two C. jejuni representative strains (NCTC 11168 and 81-176) and one C. coli strain (RM2228). Furthermore, the genomic organization of exbB-exbD-tonB loci in different Campylobacter strains was compared (Fig. S1).
Table 3.
FeEnt utilization system in representative C. jejuni and C. coli strains.
| Strain | C. jejuni | C. coli | |
|---|---|---|---|
| NCTC 11168 | 81-176 | RM2228 | |
| FeEnt utilization | Yes | No | Not determined |
| CfrA | + | − | + |
| CfrB | pseudogene | + | + |
| TonBl ExbB1/ExbD1 |
+ + |
−
− |
+ + |
| TonB2 ExbB2/ExbD2 |
+ + |
+ + |
− + |
| TonB3 ExbB3/ExbD3 |
+ + |
− + |
+ + |
| CeuBCDE | + | + | + |
| Cee | + | − | + |
| Source | human | human | Poultry |
ExbB3 and ExbD3 may not be cognate components for TonB3 because tonB3, in fact, is physically distant from the so-called exbB3/exbD3 operon. Cj0111, the gene immediately downstream of exbD3 (Cj0110) (Fig. S1), encodes a protein with a TonB-2 domain (pfam13103) at C-terminal, suggesting the ExbB3/ExbD3 system may interact with Cj0111. Interestingly, the exbB3-exbD3-Cj0111-Cj0112-Cj0113 operon resembles the genomic organization of Tol operon (tolQ-tolR-tolA-tolB-pal) identified in many other Gram-negative bacteria (Godlewska, et al., 2009) (Fig. S1).
The exbB2-exbD2-tonB2 operon is highly conserved and present in all C. jejuni genomes. Interestingly, in C. coli RM2228, tonB2 is missing while the complete exbB2/exbD2 genes are present (Table 3); this unique pattern also was observed in other sequenced C. coli strains in IMG (data not shown).
In C. jejuni NCTC 11168, a putative iron transporter gene (Cj0177) and the transferrin/lactoferrin receptor gene (Cj0178) are immediately upstream of the exbB1-exbD1-tonB1 operon (Fig. S1), suggesting that TonB1 energy transduction system is functionally related to Cj0177 and Cj0178. Cj0177 is a homolog of E. coli CjrA transporter (50% aa similarity) that is adjacent to CjrB (a TonB protein) and CjrC (an outer membrane colicin Js receptor) (Smajs & Weinstock, 2001). The TonB1 system was prevalent and highly conserved in C. coli. For example, analysis of the sequence data from a recent Campylobacter pan-genome project (Lefebure, et al., 2010) revealed that the exbB1-exbD1-tonB1 operon was present in all 42 diverse C. coli strains. However, the exbB1-exbD1-tonB1 operon was missing in 14 out of 59 C. jejuni genomes retrieved from IMG.
The TonB2 system is critical for CfrB-dependent FeEnt acquisition in C. jejuni 81-176
A library containing 7350 Tn5 mutants was screened for the mutants that failed to grow in the iron-restricted medium supplemented with Ent. Eight mutants were identified, in which five have transposons inserted in exbB2-exbD2-tonB2 locus (Table 4).
CfrB also utilizes TonB3 for efficient FeEnt acquisition
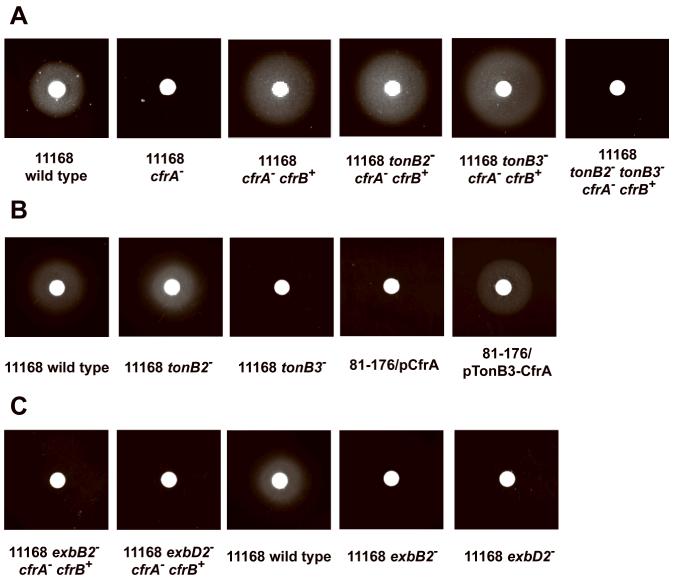
Despite the critical role of TonB2 system in CfrB-dependent pathway as demonstrated above, TonB3 may also involve CfrB-dependent FeEnt acquisition because inactivation of tonB3 in C. coli VC167, a strain using CfrB as dominant FeEnt receptor, abolished its ability to utilize Ent (Guerry, et al., 1997). Thus, we further examined the genetic interaction of CfrB with TonB3 by manipulating C. jejuni NCTC 11168. As shown in Fig. 1A, chromosomal complementation of the cfrA mutant with cfrB completely restored its ability to utilize FeEnt. Inactivation of tonB2 or tonB3 alone in this genetically repaired mutant (cfrA− cfrB+) did not affect FeEnt acquisition (Fig. 1A). However, the mutant with mutations in both tonB2 and tonB3 genes failed to utilize FeEnt, indicating CfrB also could interact with TonB3 for efficient FeEnt utilization.
Figure 1.
Identification of specific TonB-ExbB-ExbD components required for CfrA- and CfrB-dependent FeEnt acquisition in C. jejuni. Standard Ent growth promotion assay was performed for wild-type strain and its mutant derivatives. (A) TonB3 could interact with CfrB for efficient FeEnt acquisition in C. jejuni NCTC 11168. (B) TonB3 is essential for CfrA-dependent FeEnt acquisition. (C) ExbB2 and ExbD2 are essential for FeEnt acquisition in Campylobacter.
TonB3 is essential for CfrA-dependent FeEnt acquisition
Regarding the identity of specific TonB component for CfrA, in C. jejuni NCTC 11168, inactivation of TonB3 alone abolished FeEnt utilization while inactivation of TonB2 did not (Fig. 1B). In addition, for C. jejuni 81-176 that only contains TonB2 (Table 3), wild-type CfrA gene (pCfrA) failed to rescue it for FeEnt utilization while both cfrA and tonB3 genes (pTonB3-CfrA) restored its ability to utilize FeEnt (Fig. 1B); this evidence further indicated that TonB3 is critical for CfrA-dependent FeEnt acquisition.
ExbB2 and ExbD2 are essential for both CfrA- and CfrB-dependent FeEnt acquisition in C. jejuni
As revealed in random transposon mutagenesis experiment, ExbB2 and ExbD2 are essential for CfrB-dependent FeEnt acquisition in C. jejuni 81-176 background (Table 4). To further validate this finding, we transferred the exbB2 and exbD2 mutations from 81-176 mutants into the CfrB-repaired NCTC 11168 strain JL612 (cfrA− cfrB+). As shown in Fig. 1C, inactivation of ExbB2 or ExbD2 alone completely abolished JL612’s ability to utilize FeEnt, further demonstrating the essential role of ExbB2 and ExbD2 in CfrB-dependent pathway.
Regarding the CfrA-dependent pathway that requires TonB3 in wild-type NCTC 11168, single exbB2 or exbD2 mutation could completely abolish its ability to utilize FeEnt (Fig. 1C). This finding indicates that ExbB2 and ExbD2 are also essential for CfrA-dependent FeEnt acquisition even if they are distant from the tonB3-cfrA locus; other ExbB/ExbD systems in NCTC 11168, such as ExbB3/ExbD3, cannot compensate the function of ExbB2/ExbD2 for TonB3-mediated FeEnt acquisition.
Predicted TonB box in FeEnt receptors and the corresponding binding sites in TonB
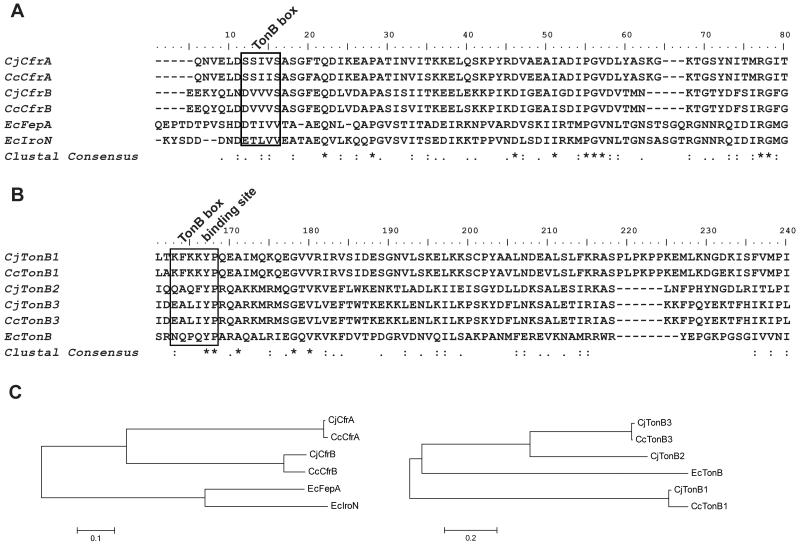
It is interesting that CfrA only interacts with TonB3 while CfrB can interact with both TonB2 and TonB3 for efficient FeEnt acquisition. Since the TonB-dependent FeEnt receptor interacts with TonB through the conserved TonB box that contains 5-7 amino acid residues (Krewulak & Vogel, 2011), we speculate the sequence variation may exist in the TonB box region of CfrA and CfrB. As expected, the predicted TonB box of CfrA is SSIV(/I)S while that of CfrB is DVVVS (Figure 2A); these specific TonB box sequences are highly conserved in the sequenced CfrA and CfrB, respectively (data not shown). Analysis of different TonB proteins indicated that the predicted TonB box binding sites for TonB1, TonB2, and TonB 3 are KFKKYP, QAQFYP, and EALIYP, respectively (Figure 2B); each specific type of TonB from diverse Campylobacter strains displays the identical TonB box binding site (data not shown).
Figure 2.
Comparative analyses of FeEnt receptors and TonB components in Campylobacter and E. coli. (A) Multiple sequence alignment of FeEnt receptors from Campylobacter and E. coli. The sequences were aligned with Clustal W2. The region of aa 1-80 was displayed. Putative TonB box region was highlighted in the rectangle boxes. Identical amino acids are marked by an asterisk (*), and conserved and semi-conserved substitutions, substitutions are marked by colon (:) and a single dot (•), respectively. (B) Multiple sequence alignment of identified TonB components in Campylobacter and E. coli. The region of aa 161-240 was displayed. One putative TonB box binding site was highlighted in the rectangle box. (C) Phylogenetic analyses of FeEnt receptors and TonB components from Campylobacter and E. coli. Left panel, phylogenetic analysis of C. jejuni FeEnt receptors (CjCfrA and CjCfrB), C. coli FeEnt receptors (CcCfrA and CcCfrB), and E. coli FepA and IroN. Right panel, phylogenetic analysis of C. jejuni TonB (CjTonB1, CjTonB2, and CjTonB3), C. coli TonB (CcTonB1 and CcTonB3), and E. coli TonB (EcTonB).
The CfrA from C. jejuni and C. coli form a cluster that is close to the cluster of the CfrB from C. jejuni and C. coli, indicating they are orthologs of the FepA and IroN from E. coli (Figure 2C, left panel). In addition, TonB2 was phylogenetically closer to TonB3 than to TonB1 (Figure 2C, right panel).
Discussion
The presence of multiple TonB-ExbB-ExbD systems in a single organism is not unique to Campylobacter although many Gram-negative bacteria, such as E. coli, only have one set of TonB-ExbB-ExbD complex. For example, Helicobacter pylori 26995 (Schauer, et al., 2007), Vibrio cholerae CA401S (Seliger, et al., 2001), and Pseudomonas aeruginosa PAO1 (Zhao & Poole, 2000) have two sets of TonB systems. In addition, there are up to eight sets of TonB-ExbB-ExbD paralogs in Myxococcus xanthus (Sogaard-Andersen, 2011). Bacteria may acquire multiple TonB systems with different functional specificities during evolution (Seliger, et al., 2001). This study is focused on the identification of cognate TonB system(s) for the two different FeEnt acquisition pathways in Campylobacter (Zeng, et al., 2013). We demonstrated that TonB3 plays a dominant role in CfrA-dependent FeEnt acquisition, which is consistent with the finding from a recent report (Naikare, et al., 2013). However, in CfrB-dependent pathway, TonB2 and TonB3 are interchangeable for efficient FeEnt acquisition.
Interestingly, although CfrA and CfrB display different specificity for TonB component, both the CfrA- and CfrB-dependent FeEnt acquisition pathways use the same ExbB/ExbD complex (ExbB2/ExbD2). Our data here suggest the Campylobacter ExbB2/ExbD2 is orthologs of the ExbB/ExbD complex in E. coli in terms of FeEnt acquisition. The pivot role of ExbB2/ExbD2 was also reflected by the presence of exbB2/exbD2 in all Campylobacter genomes. It has been proposed that ExbD interacts with TonB through the periplasmic domain (Pramanik, et al., 2011, Ollis & Postle, 2012). Thus, future structural and sequence analyses of ExbD2, TonB2 and TonB3 are needed to reveal molecular interaction between ExbD and TonB.
As expected, ExbB3/ExbD3 is not associated with TonB3 for FeEnt acquisition. Consistent with this finding, previous microarray study (Palyada, et al., 2004) has suggested that the ExbB3/ExbD3 complex is not involved in iron acquisition because ExbB3/ExbD3 genes were up-regulated in response to iron repletion rather than iron depletion. The exbB3, exbD3, and their downstream genes display similar genomic organization to the tolQ-tolR-tolA-tolB-pal operon (Fig. S1) that is important in the pathophysiology of other Gram-negative bacteria (Godlewska, et al., 2009). This observation warrants further functional characterization of the ExbB3/ExbD3 in Campylobacter.
Current structural model for the interaction between TonB and TonB-dependent receptor is the formation of the β-sheet containing mixed β-strands from TonB and TonB-dependent receptor (also called TonB box) (Shultis, et al., 2006). In this study, we observed that CfrA and CfrB displayed different TonB box sequence with major difference in the first two aa residues. The first two residues of the CfrA TonB box (SS) contain uncharged polar side chain. However, the first two residues of the CfrB TonB box (DV) contain acidic side chain and nonpolar side chain, respectively. Thus, the first two aa residues in the TonB box of CfrA or CfrB may determine its specificity for the cognate TonB component. This hypothesis needs to be examined in the future.
Supplementary Material
Acknowledgements
We thank Dr. Richard D. Haigh (University of Leicester, UK) for providing cfrA::erm mutant DNA, Drs. Ky Van Hoang and Zhong Wang for technical support, Ms. Barbara Gillespie for careful proofreading of this manuscript. This work was supported by NIH grant 1R56AI090095-01A1 and Agriculture and Food Research Initiative (AFRI) Competitive Grant No. 2012-68003-19679.
References
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157:472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- Braun V, Hantke K, Koster W. Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst. 1998;35:67–145. [PubMed] [Google Scholar]
- Chaudhuri RR, Loman NJ, Snyder LA, Bailey CM, Stekel DJ, Pallen MJ. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 2008;36:D543–546. doi: 10.1093/nar/gkm928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE, Mongodin EF, Mandrell RE, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewska R, Wisniewska K, Pietras Z, Jagusztyn-Krynicka EK. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol Lett. 2009;298:1–11. doi: 10.1111/j.1574-6968.2009.01659.x. [DOI] [PubMed] [Google Scholar]
- Guerry P, Perez-Casal J, Yao R, McVeigh A, Trust TJ. A genetic locus involved in iron utilization unique to some Campylobacter strains. J Bacteriol. 1997;179:3997–4002. doi: 10.1128/jb.179.12.3997-4002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang KV, Wang Y, Lin J. Identification of genetic loci that contribute to Campylobacter resistance to fowlicidin-1, a chicken host defense peptide. Front Cell Infect Microbiol. 2012;2:32. doi: 10.3389/fcimb.2012.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang KV, Stern NJ, Saxton AM, Xu F, Zeng X, Lin J. Prevalence, development, and molecular mechanisms of bacteriocin resistance in Campylobacter. Appl Environ Microbiol. 2011;77:2309–2316. doi: 10.1128/AEM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreuter D, Tsai J, Watson RO, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon B, Wang Y, Hao H, Barton YW, Zhang Q. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J Antimicrob Chemother. 2011;66:79–85. doi: 10.1093/jac/dkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev AV, Wren BW. Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl Environ Microbiol. 2005;71:4004–4013. doi: 10.1128/AEM.71.7.4004-4013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewulak KD, Vogel HJ. TonB or not TonB: is that the question? Biochem Cell Biol. 2011;89:87–97. doi: 10.1139/o10-141. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebure T, Bitar PD, Suzuki H, Stanhope MJ. Evolutionary dynamics of complete Campylobacter pan-genomes and the bacterial species concept. Genome Biol Evol. 2010;2:646–655. doi: 10.1093/gbe/evq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wang Y, Hoang KV. Systematic identification of genetic loci required for polymyxin resistance in Campylobacter jejuni using an efficient in vivo transposon mutagenesis system. Foodborne Pathog Dis. 2009;6:173–185. doi: 10.1089/fpd.2008.0177. [DOI] [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CE, Williams PH, Ketley JM. Pumping iron: mechanisms for iron uptake by Campylobacter. Microbiology. 2009;155:3157–3165. doi: 10.1099/mic.0.032425-0. [DOI] [PubMed] [Google Scholar]
- Naikare H, Butcher J, Flint A, Xu J, Raymond KN, Stintzi A. Campylobacter jejuni ferric-enterobactin receptor CfrA is TonB3 dependent and mediates iron acquisition from structurally different catechol siderophores. Metallomics. 2013 doi: 10.1039/c3mt20254b. May 24 Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis AA, Postle K. Identification of functionally important TonB-ExbD periplasmic domain interactions in vivo. J Bacteriol. 2012;194:3078–3087. doi: 10.1128/JB.00018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palyada K, Threadgill D, Stintzi A. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol. 2004;186:4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Pi H, Jones SA, Mercer LE, et al. Role of catecholate siderophores in gram-negative bacterial colonization of the mouse gut. PLoS One. 2012;7:e50020. doi: 10.1371/journal.pone.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik A, Hauf W, Hoffmann J, Cernescu M, Brutschy B, Braun V. Oligomeric structure of ExbB and ExbB-ExbD isolated from Escherichia coli as revealed by LILBID mass spectrometry. Biochemistry. 2011;50:8950–8956. doi: 10.1021/bi2008195. [DOI] [PubMed] [Google Scholar]
- Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K, Gouget B, Carriere M, Labigne A, de Reuse H. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol Microbiol. 2007;63:1054–1068. doi: 10.1111/j.1365-2958.2006.05578.x. [DOI] [PubMed] [Google Scholar]
- Seliger SS, Mey AR, Valle AM, Payne SM. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol Microbiol. 2001;39:801–812. doi: 10.1046/j.1365-2958.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- Shultis DD, Purdy MD, Banchs CN, Wiener MC. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- Smajs D, Weinstock GM. The iron- and temperature-regulated cjrBC genes of Shigella and enteroinvasive Escherichia coli strains code for colicin Js uptake. J Bacteriol. 2001;183:3958–3966. doi: 10.1128/JB.183.13.3958-3966.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard-Andersen L. Directional intracellular trafficking in bacteria. Proc Natl Acad Sci U S A. 2011;108:7283–7284. doi: 10.1073/pnas.1104616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Vliet AHMV, Ketley JM. Iron Metabolism, Transport, and Regulation. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. ASM Press; Washington, DC: 2008. pp. 591–610. [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Wang Y, Taylor DE. Natural transformation in Campylobacter species. J Bacteriol. 1990;172:949–955. doi: 10.1128/jb.172.2.949-955.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Taylor DE. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- Xu F, Zeng X, Haigh RD, Ketley JM, Lin J. Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter. J Bacteriol. 2010;192:4425–4435. doi: 10.1128/JB.00478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Alm RA, Trust TJ, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- Zeng X, Xu F, Lin J. Molecular, antigenic, and functional characteristics of ferric enterobactin receptor CfrA in Campylobacter jejuni. Infect Immun. 2009;77:5437–5448. doi: 10.1128/IAI.00666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Mo Y, Xu F, Lin J. Identification and characterization of a periplasmic trilactone esterase, Cee, revealed unique features of ferric enterobactin acquisition in Campylobacter. Mol Microbiol. 2013;87:594–608. doi: 10.1111/mmi.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Poole K. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol Lett. 2000;184:127–132. doi: 10.1111/j.1574-6968.2000.tb09002.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.