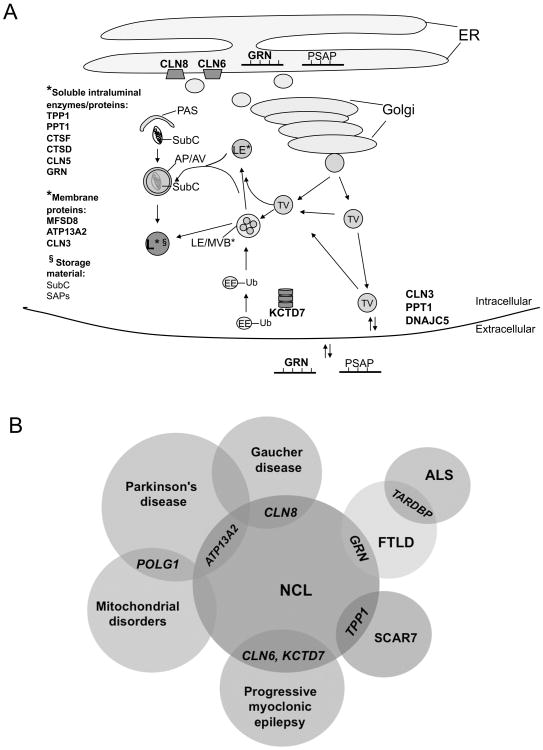
Figure 1. The broadening NCL biological pathway and genotypic and phenotypic spectrum.
A. A hypothetical model of the expanding NCL biological pathway(s) is depicted. The genes mutated in NCL patients are indicated by boldface type and their putative protein cellular localizations are shown (*). The NCL proteins may function in common or intersecting pathways. Alternatively, one or more NCL proteins may function in parallel pathways that, when dysfunctional, lead to a shared pathology. Progranulin (GRN) and prosaposin (PSAP) similarly exist as both precursor polypeptides in the endoplasmic reticulum (ER) and in the secretory pathway (Golgi and transport vesicles [TV]) and extracellular space, and are cleaved into lower molecular weight peptides in the endosomal-lysosomal system. Lines on GRN and PSAP depict cleavage sites. GRN, PSAP, and cathepsin D (CTSD) are sorted within the secretory pathway and endosomal system via sortilin. CLN5 also binds sortilin/retromer complex. Ubiquitination (Ub) is an increasingly recognized mechanism for regulating trafficking in the endosomal-lysosomal system. Subunit c (SubC) and the saposins (SAPs), cleaved from PSAP, are the major proteins accumulating in NCL storage material within autophagic vacuoles (AV) and lysosomes (L). Subunit c is the pore-forming subunit of the mitochondrial ATP synthase complex (Complex V), and the SAPs are lipid carrying accessory proteins in the endosomal-lysosomal system. *indicates the protein localization; §indicates localization of the listed storage material. TV=transport vesicle; EE=early endosome; Ub=ubiquitin; LE=late endosome; MVB=multivesicular body; PAS=preautophagosomal structure; AP/AV=amphisome/autophagic vacuole; L=lysosome. B. Venn diagram showing that new NCL genes (GRN, ATP13A2, and KCTD7) as well as previously defined NCL genes (TPP1, CLN6, and CLN8) genes intersect with other neurodegenerative disorders. As described in the text, polymorphisms in the CLN8 locus that influence its expression appear to be genetic modifiers of Gaucher disease. Other genes such as POLG and TARDBP, although not specifically identified as causative genes in NCL, may offer additional insight into NCL biology given their association with diseases showing clinicopathological overlap with NCL, including Parkinson's disease, mitochondrial disease, and frontotemporal lobar dementia (FTLD). TARDBP (TAR DNA-binding protein 43) encodes TDP-43, the protein that loses its nuclear localization and forms ubiquitinated, hyperphosphorylated inclusions in various forms of FTLD and amyotrophic lateral sclerosis (ALS). SCAR7, autosomal recessive spinocerebellar ataxia type 7.