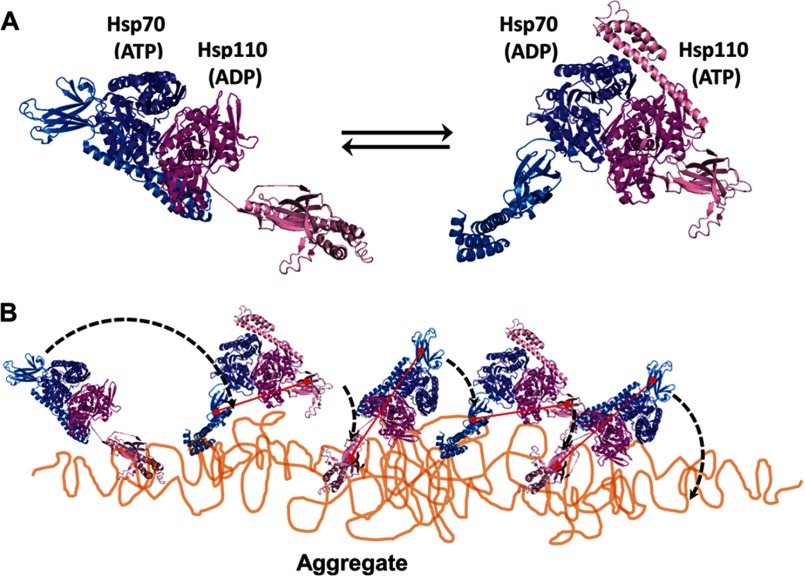
FIGURE 7.
Model for Hsp110-Hsp70-mediated disaggregation. A, model of the two alternating functional HSPA8-SSE1 heterodimers, respectively, in the ATP-ADP state (left panel) and the ADP-ATP state (right panel), reconstructed by structural alignments of the SBDs from HSPA8 (blue tones) and SSE1 (magenta tones), either with the closed ADP-bound form of DnaK (PDB code 2KHO) or with the opened ATP-bound form of DnaK (PDB code 4B9Q). The association of the HSPA8 NBD with the SSE1 NBD was from PDB code 3C7N. The substrate binding sites of HSPA8 and SSE1 SBDs are shown as red circles. The connecting red line shows the distances between binding sites in the heterodimer. B, model for the disaggregation mechanism. The crystal structure of the HSPA8-SSE1 heterodimer (32) suggests that when one chaperone is ADP-bound, its SBD can be locked on an unfolded substrate, whereas the other chaperone is in the ATP-bound state with a widely opened SBD. During disaggregation, SSE1 and Hsp70 may thus alternate between a closed locked state, anchoring the whole chaperone heterodimer to the aggregate surface at a single misfolded loop (left panel), becoming unfolded by entropic pulling (44), and an opened state that can scan far from the anchor for new misfolded structures to bind to. Then, upon ATP hydrolysis, the second chaperone may bind and apply an unfolding pressure on the newly targeted misfolded loops (right panel). When successful, locking may unfold the misfolded structure, while concomitantly, an allosteric signal is sent from the NBD of the second locking chaperone to the NBD of the first chaperone to release ADP and thus unlock the SBD from a now newly enlarged unfolded anchoring loop, which may then refold to the native state.