Figure 4. Comparison of the R2 ribozyme from 5 silkmoth species.
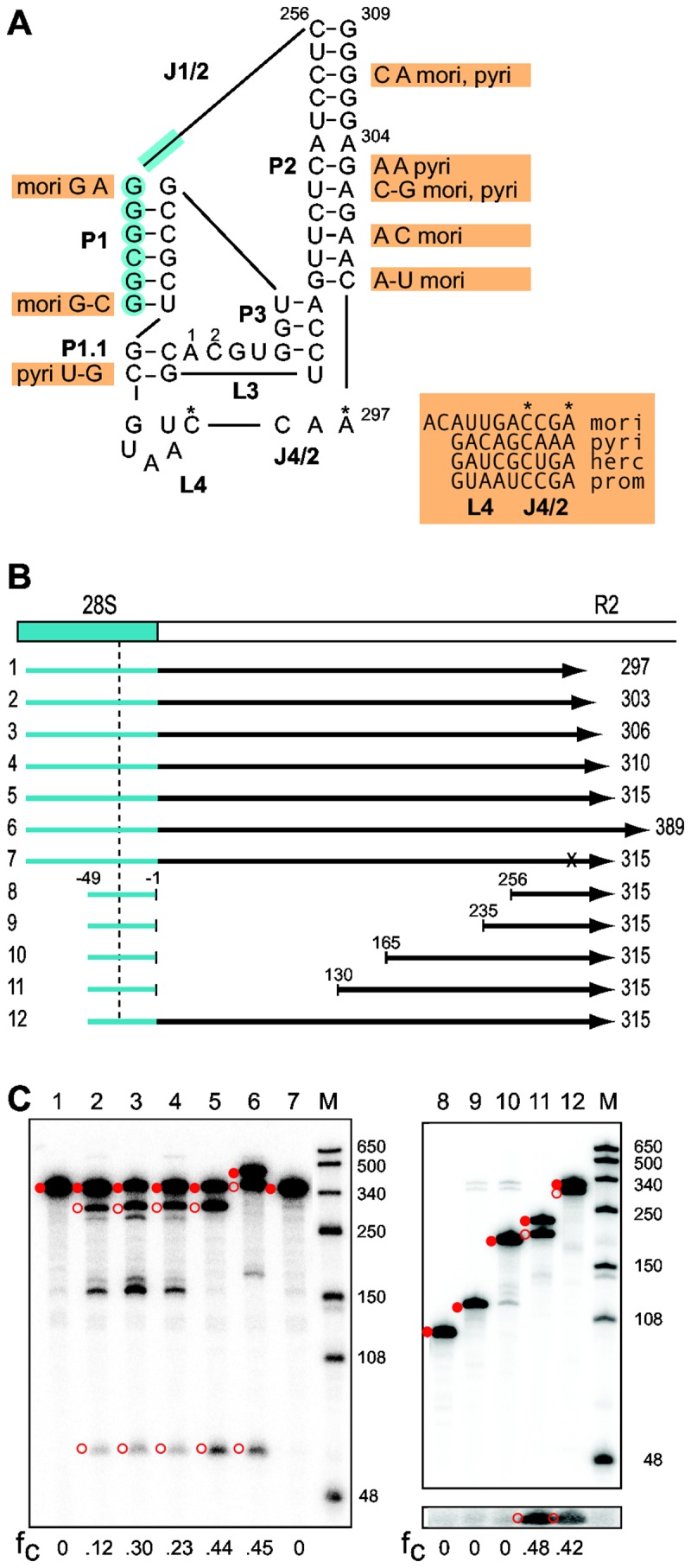
(A) Secondary structure of the putative ribozyme encoded by the R2 element of S . cynthia . Blue shading indicates nucleotides from the 28S gene. Indicated next to the S . cynthia sequence are differences in the R2 sequences from 4 silkmoth species (nucleotides in orange boxes). The entire sequence of the largely unconserved L4- J4/2 region is presented for each species. Two species contained nucleotide differences in the otherwise conserved L3 (residue 1 is a 'U' in S . pyri , residue 2 is an 'A' in C . hercules ). The J1/2 loop length varied from 216 nt in S . pyri to 498 nt in C . hercules mori, B. mori; herc, C . hercules ; prom, C . promethea ; pyri, S . pyri . (B) Diagrams of the 28S/R2 RNAs from S . cynthia tested for self-cleavage as described in Figure 2. The vertical dashed line indicates the predicted upstream cleavage site at -28. Negative numbers indicate position in the 28S gene relative to the R2 insertion site; positive numbers indicate position in the S . cynthia R2. The x in RNA 7 represents a 'U' substitution for the putative catalytic 'C'. Templates 8-11 have varying amounts of the J1/2 loop removed. (C) 5% denaturing acrylamide gels showing the products from the co-transcription/self-cleavage assays. Lane numbers correspond to the RNAs in panel (B). The uncleaved RNA (solid red circle) and self-cleavage products (open red circles) are indicated. Lane M, RNA length markers with sizes indicated. For RNAs 8-12, a longer exposure of the lower portion of the gel is shown to better visualize the 21 nt upstream cleavage products. The fraction of the synthesized RNA undergoing self-cleavage (fc) is shown at the bottom of each gel.
