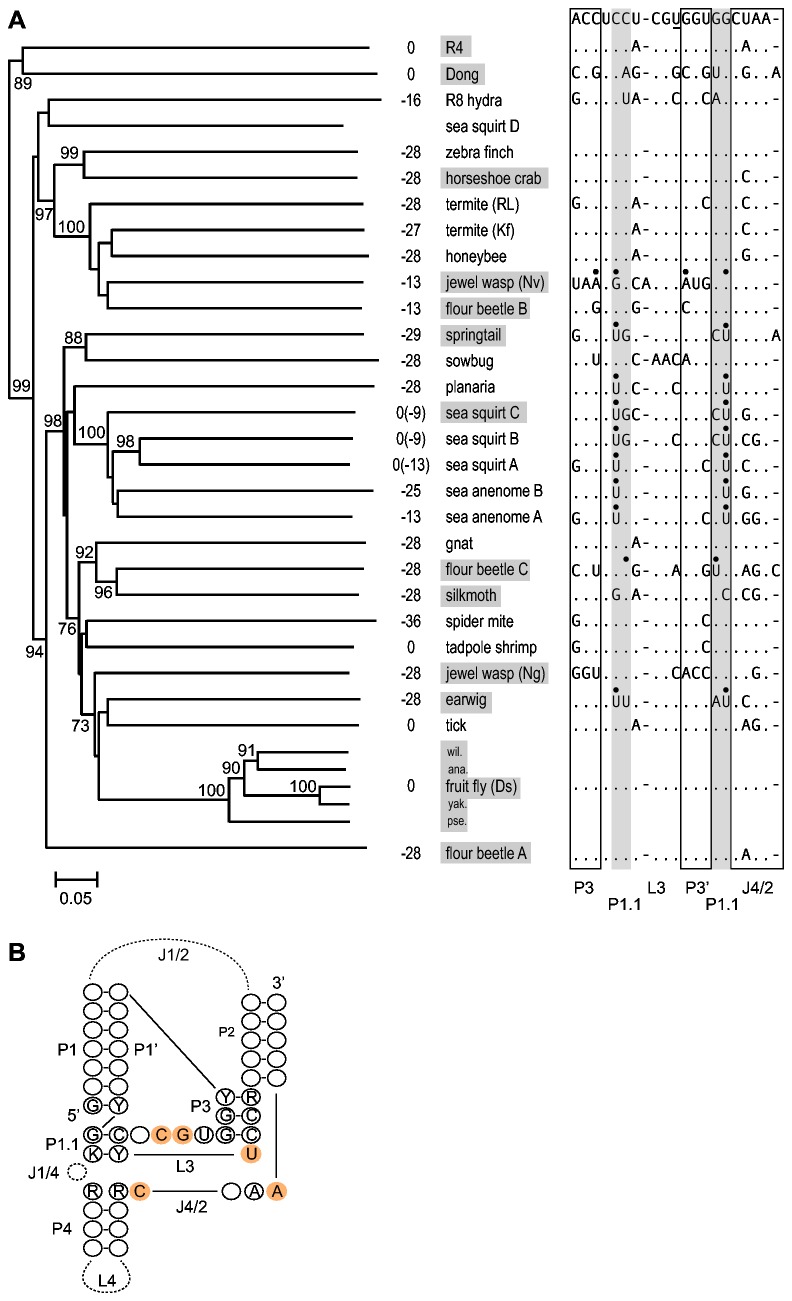
Figure 6. R2 element phylogeny and its correlation with cleavage location and ribozyme sequence.
(A) Best tree based on the neighbor-joining method and rooted using the retrotransposons Baggins1 and L1Tc. Most elements are listed in Figure 2 and Supporting Information, Figure S2. Additional fruit fly elements include: D . willistoni (wil.), D . ananassae (ana.), D . yakuba (yak.), and D . pseudoobscura (pse.). Bootstrap values above 70% are shown. Numbers left of the elements are predicted or demonstrated (boxed elements) self-cleavage locations relative to the R2 insertion site. The 5' junction cleavage site agrees with previous predictions for sea squirt A, tick, and R4; however, the tadpole 5' end is shifted 1 nucleotide and 28S cleavage sites are suggested for zebra finch, gnat, and both termites [12]. The 5' end of the sea squirt D element could not be folded and may be incomplete. To the right of each element are sequence differences in the ribozyme active site relative to the R2 consensus sequence (top line). The underlined 'U' in this consensus is a 'C' in the HDV antigenomic ribozyme. Black dots indicate nucleotides that disrupt P3 or P1.1 stem pairing. (B) Consensus structure of the R2 ribozymes based on 26 elements (additional fruit fly elements were not included) in panel (A). Length differences and mismatches in the P1, P2, and P3 stems are described in the text. Nucleotides present in at least 75% of the R2 ribozymes are indicated. Invariant nucleotides are shaded orange. The sow bug ribozyme, which does not show activity in vitro, has 'C' to 'A' and 'G' to 'A' differences at the conserved sites in L3. Y= U or C; R= A or G; K= G or U.