Fig. 5.
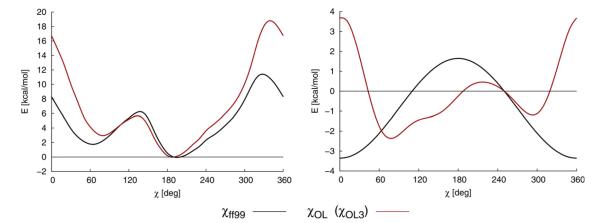
Example of dihedral profiles in MM. Complete MM torsion profile (i.e., all the force field terms) for the χ angle (on the left) calculated in vacuum and the net χ dihedral term (on the right) of ff99 or parmbsc0 (black) and parmχOL3 (red). Data for model of RNA cytidine are shown [121]. Note that the “Olomouc” force field is a major reparameterization. The modest difference in the slope in the anti to high anti region ( ~190°–280°) is critical to prevent degradation of RNA simulations to structures with DNA-like high-anti χ. (ParmχOL3 and parmχOL are abbreviations of the same force field, the dual name is due to historical reasons) [120,121]. Although the “toxicity” of the high-anti bias of the original force field versions for G-RNA has not been tested, we advise to use the parmbsc0 + parmχOL3 force field combination for longer G-RNA simulations. The G-RNA loop regions are definitely going to profit from applying the parmχOL3.
