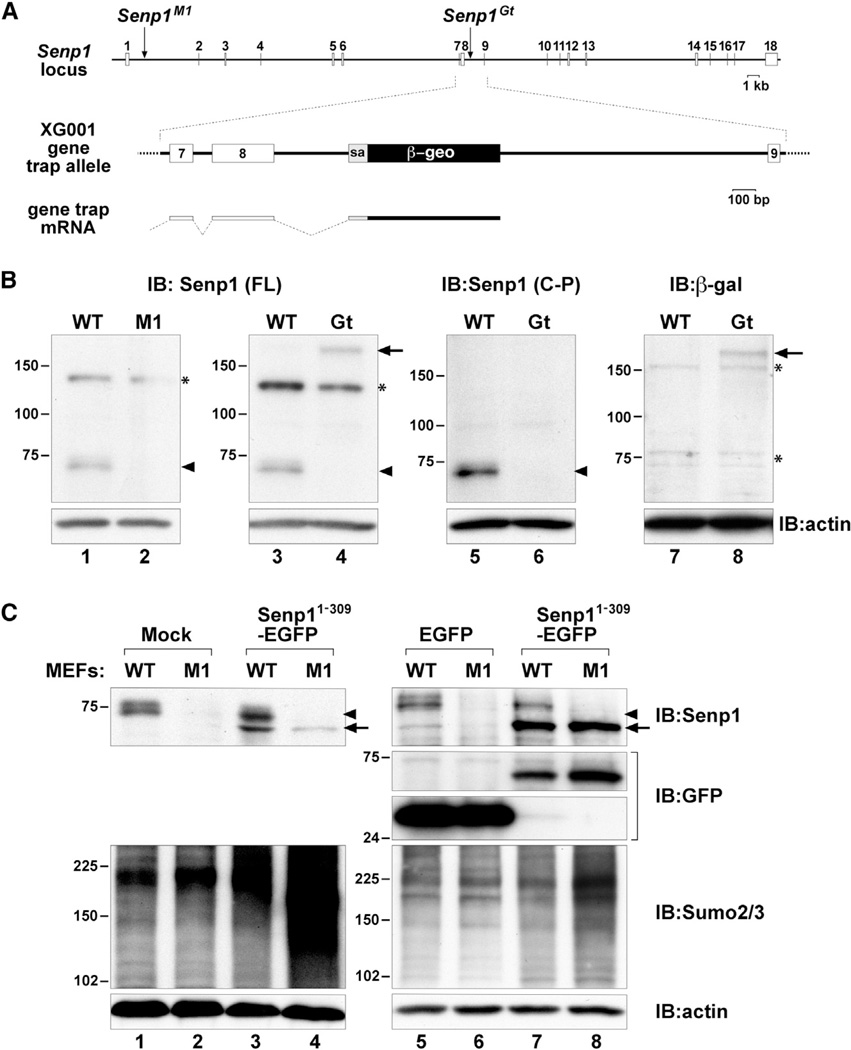
Figure 3. Structure, Expression, and Interfering Activity of the SenpGt Allele.
(A) Schematic representation of the entire Senp1 locus showing the M1 retroviral vector insertion in intron 1 and the gene trap vector insertion in intron 8. A close-up of the structure of the XG001 gene trap insertion is shown in the middle. Shown below is the fusion transcript resulting from splicing from exon 8 to the splice acceptor (sa) of the β-geo cassette.
(B) Immunoblots of lysates of WT, M1, or Gt embryos detected with anti-Senp1 raised against full-length (FL) protein (lanes 1–4) or against a carboxy-terminal peptide (C-P) (lanes 5 and 6), or anti-β-gal (lanes 7 and 8). The 73 kDa Senp1 protein (arrowheads) is not detected ineither mutant. The 180 kDa band (arrows) detected with both anti-Senp1 (FL) and anti-β-gal represents Senp1 exons 1–8 fused to β-geo. Asterisks (*) mark nonspecific bands.
(C) Immunoblots of lysates of WT or M1MEFs either mock infected (lanes 1 and 2), or infected with Senp11–309-EGFP retroviral vector (lanes 3 and 4, 7 and 8) or control EGFP-only vector (lanes 5 and 6). Top panels show detection with anti-Senp1. Arrowheads indicate endogenous Senp1 protein, and arrows indicate the Senp11–309-EGFP fusion protein. Lanes 5–8 in the upper-middle panels show detection with anti-GFP. Lower-middle panels show detection with anti-Sumo2/3. Expression of Senp11–309-EGFP results in significantly higher Sumo2/3 steady-state levels only in M1 MEFs (lanes 4 and 8). Bottom panels show detection of anti-actin to control for equal loading.
See also Figure S3.