Figure 2. Building a structural model of the Sin synaptosome.
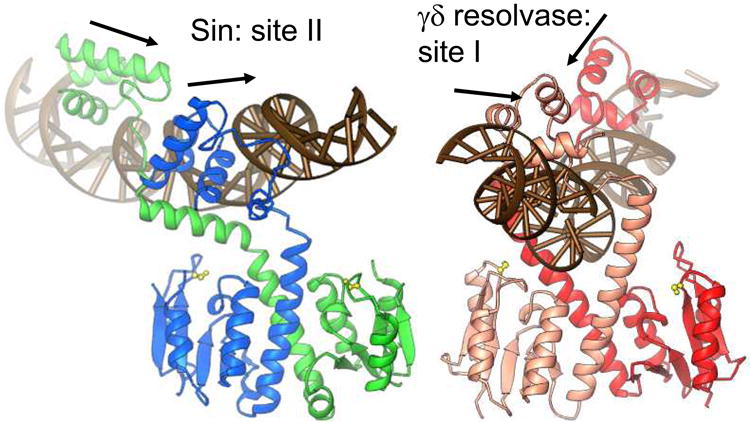
a) Structure of the Sin – site II complex (left), and comparison to the WT γδ resolvase – site I complex (right) [5, 16]. Arrows show the directions of individual half site sequences, which form a direct repeat for Sin site II and an inverted repeat for site I. Active site serines are shown as yellow balls and sticks.
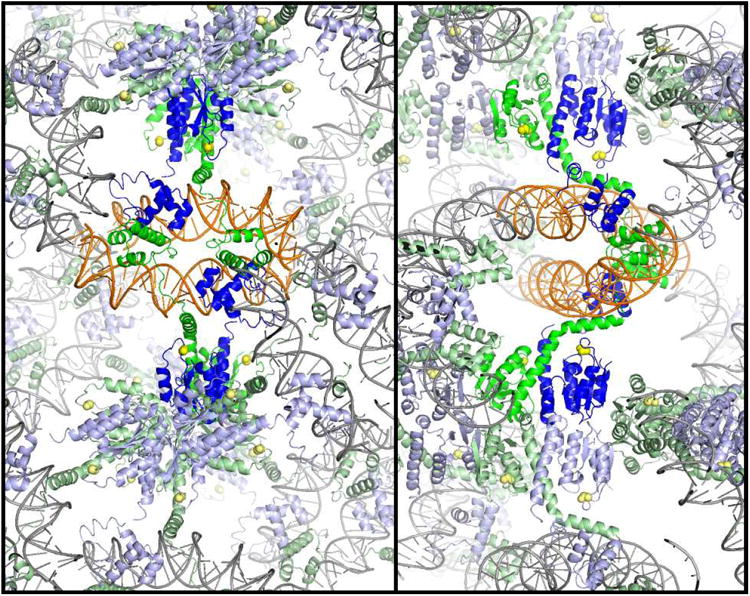
b) Packing of the Sin – Site II complex. Two orthogonal cross-sections of the crystal lattice are shown, viewed down the z axis. Individual Sin dimer – site II complexes are colored as in figure 1a, with two dimers representing a biologically relevant tetramer shown in brighter colors. The crystal contacts include end-to-end stacking of the DNA duplexes, contacts between DNA binding domains, and contacts between catalytic domains.
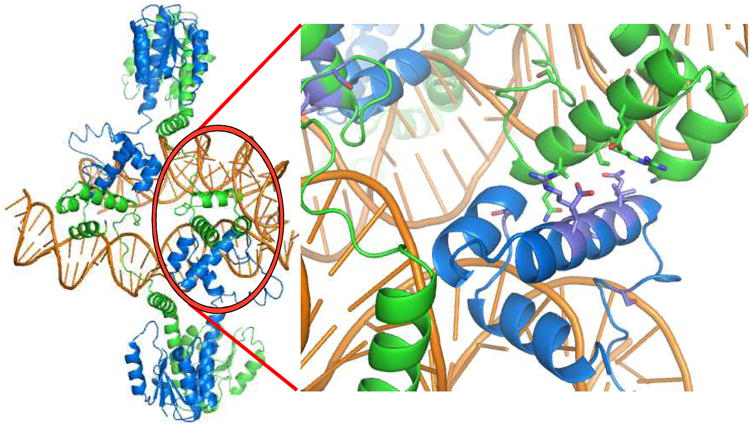
c) The biologically relevant tetramer (left) is found within the crystal, and is mediated by contacts between DNA binding domains. Residues whose mutation interferes with site II – site II contacts and recombination in vivo are shown in the closeup at right.
