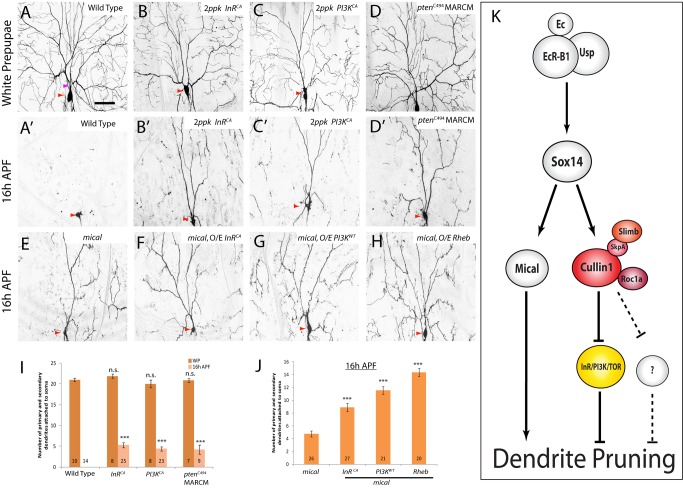
Figure 9. Activation of the InR/PI3K/TOR pathway alone is sufficient to inhibit ddaC dendrite pruning.
(A–H) Live confocal images of ddaC neurons expressing UAS-mCD8-GFP driven by ppk-Gal4 at WP or 16 h APF. Red arrowheads point to the ddaC somas. Activation of the InR/PI3K/TOR pathway through overexpression of InRCA (B, B′), PI3KCA (C, C′), or ptenc494 MARCM (D, D′) led to prominent pruning defects at 16 h APF. While mical ddaC neurons displayed an obvious pruning defect at 16 h APF (E), activation of the InR/PI3K/TOR pathway by InRCA (F), PI3KWT (G), or Rheb (H) significantly enhanced the pruning defects in the mical ddaC neurons. (I and J) Quantification of the average number of primary and secondary dendrites attached to the soma of mutant ddaC neurons at WP and 16 h APF. The number of samples (n) in each group is shown on the bars. Error bars represent S.E.M. Dorsal is up in all images. ***p<0.001, n.s., not significant. The scale bar is 50 µm. See genotypes in Text S1. (K) A model for the Cul1-based SCF E3 ligase and the InR/PI3K/TOR pathway during ddaC dendrite pruning. The Cul1-based SCF E3 ligase acts downstream of EcR-B1 and Sox14 but in parallel to the Mical pathway during ddaC dendrite pruning. The Cul1-based SCF E3 ligase complex facilitates ddaC dendrite pruning primarily through inactivation of the InR/PI3K/TOR pathway. The InR/PI3K/TOR pathway negatively regulates ddaC dendrite pruning.