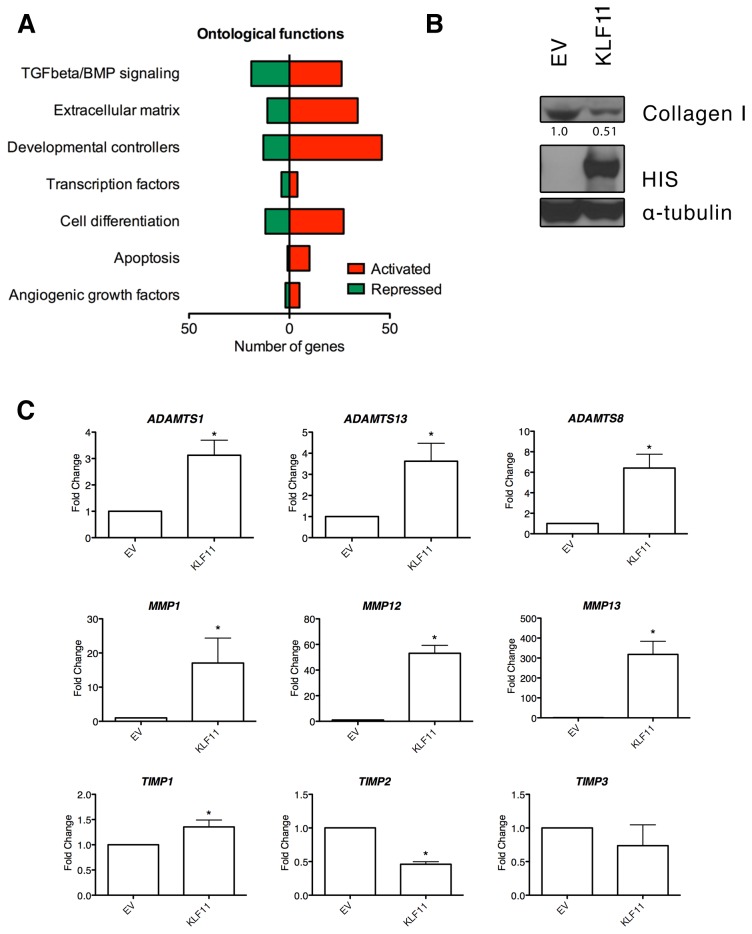
Figure 1. KLF11 is critical to a variety of biological processes in liver mesenchymal cells.
(A) LX2 cells overexpressing KLF11 elicit a variety of changes in extracellular matrix, TGFβ response, and growth factor genes. The biosynthetic response of 232 unique genes was measured by qPCR arrays (SA Biosciences) in LX2 hepatic stellate cells comparing empty vector (EV) and KLF11 transduced cells after 48 hours. The full list of altered genes may be found in Table S1. The number of genes significantly regulated by KLF11 compared to empty vector control across a variety of ontological categories is represented here. (B) Protein levels of collagen are decreased in response to KLF11 overexpression in LX2 cells. Collagen I levels were measured by western blot in LX2 hepatic stellate cells comparing EV and KLF11 transduced cells after 48 hours of infection. In the same samples, overexpression of His-KLF11 was confirmed by Omni-D8 western blot and α-tubulin confirms equal loading of lysates. Relative densitometry of collagen I levels is shown, normalized to α-tubulin control for each sample. (C) Transcript levels of matrix metalloproteinases are greatly induced while their regulators, TIMPs, are generally unchanged or downregulated. For KLF11 overexpression compared to empty vector: ADAMTS1, 3.12 ± 0.57; ADAMST13, 3.62 ± 0.85; ADAMTS8, 6.41 ±1.35; MMP1, 17.07 ± 7.30; MMP12, 53.08 ± 6.24; MMP13, 318.10 ± 65.98; TIMP1, 1.35 ± 0.14; TIMP2, 0.46 ± 0.04; TIMP3, 0.74 ± 0.31. * p-value <0.05.