Figure 5. Application of source diagnostic δ13CEAA patterns in food web studies across three different ecosystems.
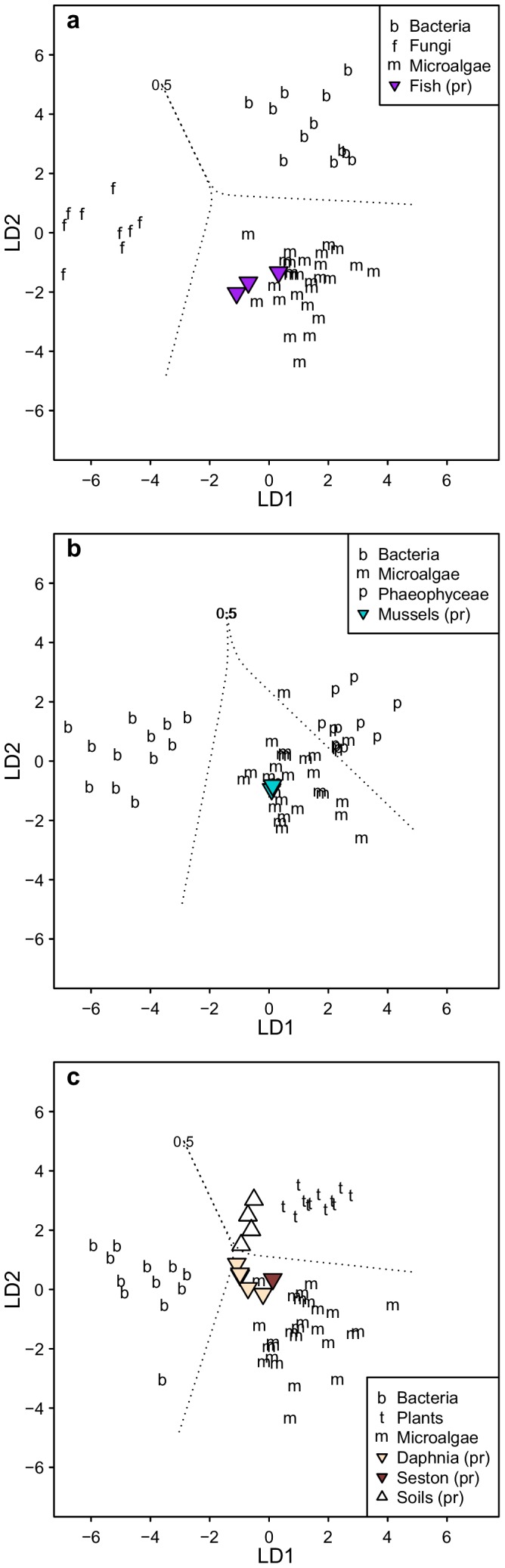
(a) In oligotrophic arctic lakes in Alaska, Daphinia sp. and seston cluster closely to each other, and their EAAs appear to derive predominantly from microalgae although a part of their EAAs may have come from foods reworked by bacteria or from allochtonous sources (i.e soils). (b) In the central North Pacific Ocean the EAAs of the carnivorous fish species (opah; Lampris guttatus, common dolphinfish; Coryphaena hippurus, broadbill swordfish; Xiphias gladius) resembled microalgae rather than EAAs from bacteria and fungi. (c) In a complex littoral marine system by the Californian shore, the δ13CEAA fingerprints of California mussel (Mytilus californianus) resemble microalgae and not bacteria or brown algae, i.e. kelp. In the figure legend, ‘Pr’ signifies predicted samples. See Table S8 for analytical details.
