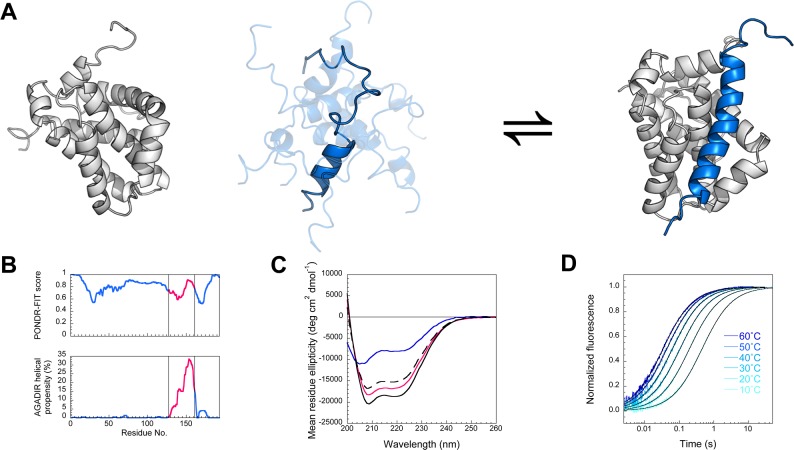
Figure 1.
(A) Cartoon depicting Mcl-1 (gray) binding PUMA peptide (blue). Unbound Mcl-1 is based on pdb 1WSX, ensemble of structures of unbound PUMA peptide built using Chimera (UCSF)22 and bound structure is based on pdb 2ROC. Figure prepared using PyMol. (B) Full-length PUMA is predicted to be entirely disordered, producing a PONDR-FIT23 score 0.5–1.0, and has residual helicity only in the BH3 region used in this study (magenta), as predicted by the helical propensity predictor AGADIR.24 (C) Consistent with a coupled folding and binding reaction, PUMA peptide binds Mcl-1 with an increase in helicity, as shown by circular dichroism. The 1:1 complex (black solid line) has a greater α-helical signal than the spectrum predicted for no interaction (dashed line), which is the sum of the PUMA alone (blue) and Mcl-1 alone (magenta) spectra. (D) Kinetics of association between Mcl-1 and PUMA peptide could be followed by stopped-flow fluorescence. An increase in temperature accelerates association. Fits for irreversible association (eq 3) are shown as black lines.