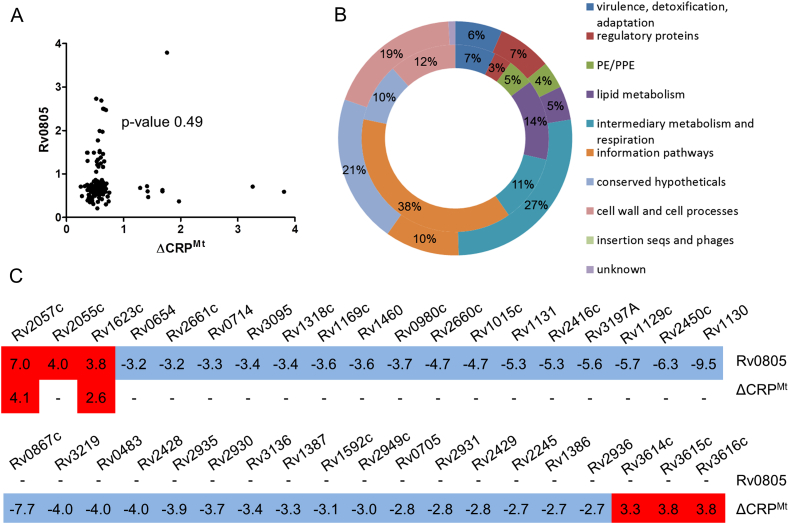
Figure 2.
Comparison of changes in the transcriptome following Rv0805 overexpression and in the Δcrp mutant A. Comparison of Rv0805 overexpression gene set and the CRPMt regulon.20,22 Fold regulation of 160 genes from the two data sets were compared on a scatter plot. Pearson's correlation analysis was performed using Graphpad Prism and was found not to be statistically significant (p-value 0.49). B. Comparison of cellular pathways affected by Rv0805 overexpression (outer donut) and CRPMt deletion (inner donut). Genes with altered expression levels (>±2 fold) were assigned functional categories according to Tuberculist (www.tuberculist.epfl.ch) annotation. Numbers represent percentage of total genes altered. C. Comparison of genes altered by Rv0805 overexpression and CRPMt deletion. Genes dysregulated upon Rv0805 overexpression (upper panel) do not show significant alteration in expression levels in the CRPMt knockout and vice-versa (lower panel). Red boxes indicate up-regulation and blue boxes indicate down-regulation compared to appropriate reference strains. Numbers indicate fold change in expression level as obtained from microarray analyses. ‘-’ indicates no change in expression levels (<2.0 fold).