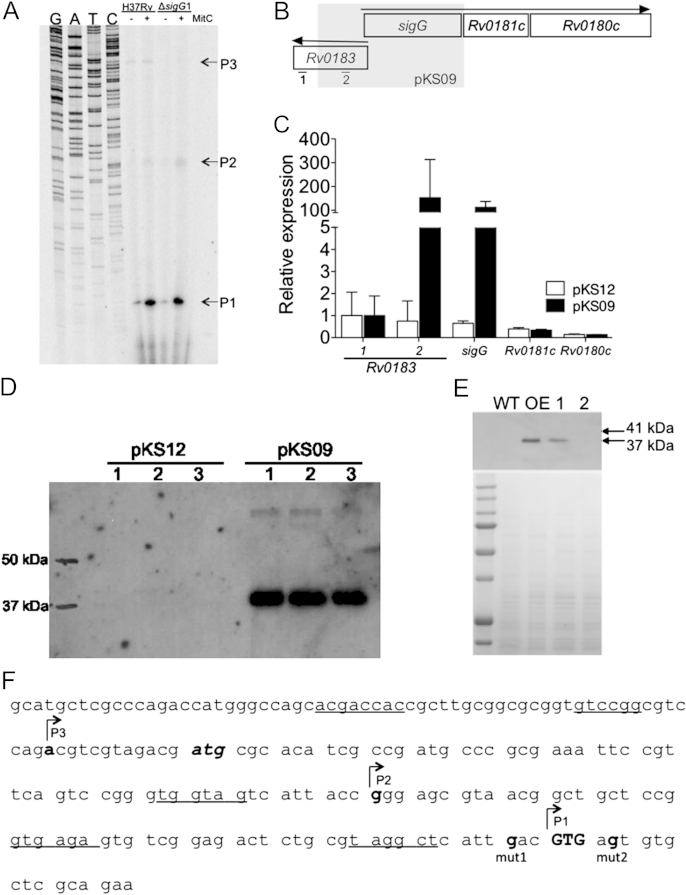
Figure 2.
Transcriptional and translational analysis of sigG. (A) Primer extension analysis using primer sigGPExt, located within the sigG annotated coding region. RNA was isolated from wild-type M. tuberculosis H37Rv and ΔsigG1 with and without treatment with 0.02 μg ml−1 mitomycin C (MitC) for 16 h. (B) Schematic of sigG operon and Rv0183 gene organisation showing region contained within pKS09 (SigG over expression construct, highlighted in grey) and position of Rv0183 primers used in (C). (C) qRT-PCR showing expression of sigG, Rv0181c, Rv0180c and Rv0183 from primers located outside (1) or within (2) pKS09 in wild-type H37Rv containing the SigG over-expression construct (pKS09) or empty vector (pKS12). Data shows expression normalised to sigA, mean + standard deviation for 3 biological replicates. (D) Western blot analysis using anti-SigG antibody of three colonies of wild-type H37Rv containing the SigG over-expression construct (pKS09) or empty vector (pKS12). Sizes of marker bands are indicated. (E) Translational start site assay. Western blot using anti-SigG antibody against cell free extracts from M. smegmatis containing no vector (WT), or M. smegmatis containing pKS09 SigG over-expression construct (OE), pKS09TTSmut1 (1) or pKS09TTSmut2 (2) translational start site frame shift mutations of SigG, deletions as indicated in (F). Equal loading was verified by Coomassie Blue staining of a similar gel (shown below Western blot). The expected sizes for SigG starting at the annotated start site (41 kDa), or the experimentally determined start site (37 kDa) are indicated. (F) DNA sequence of promoter region of sigG showing position of transcriptional start sites identified in (A), P1, P2 and P3 with potential promoter regions underlined, annotated start codon bold italics, experimentally determined start codon BOLD CAPITALS, and location of single base pair deletions used in (D) mut1 and mut2.