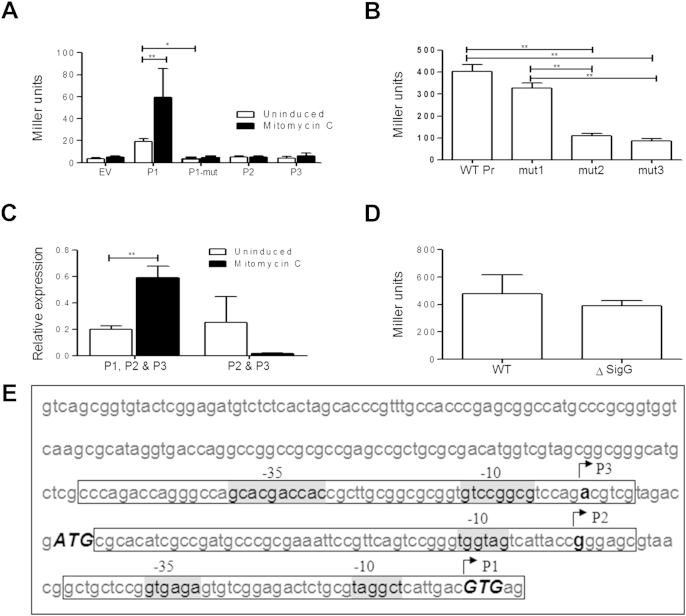
Figure 3.
Promoter activity for sigG. (A) β-galactosidase activity for M. tuberculosis containing pEJ414 empty vector (EV), or vectors containing translational fusions of each promoter identified in Figure 2A to reporter gene lacZ (as indicated in E), pLDlac1 (P1), pLDlac2 (P2) and pLDlac3 (P3) and P1 containing a mutation of the −10 region (pLDlac1-mut, P1-mut) with (black bars) and without (white bars) treatment with 0.02 μg ml−1 mitomycin C. (B) Translational fusion of 339 bp region upstream of sigG containing all 3 putative promoters (pAG04, WT Pr) with mutations to −10 regions of P1 (pAG04-mut1), P1 and P2 (pAG04-mut2) and P1, P2 and P3 (pAG04-mut3). (C) qRT-PCR using primers sigGqRTF and sigGqRTR (located downstream from P1, detecting transcript from all promoters), and sigGP2qRTF and sigGP2qRTR (located between transcriptional starts sites P1 and P2, detecting transcript starting upstream of P1). Expression was assessed from wild-type M. tuberculosis H37Rv with (black bars) and without (white bars) mitomycin C treatment and normalised to sigA. (D) β-galactosidase activity for pAG04, containing the full length sigG promoter in wild-type M. tuberculosis H37Rv and ΔsigGWO. (E) 339 bp DNA sequence of promoter region of sigG used to make construct pAG04 including P1, P2 and P3 with potential −10 and −35 promoter regions shaded grey. Locations of the DNA fragments used in the individual pLDlac promoter constructs are indicated by boxes, together with the transcriptional start site for each promoter (arrow and bold) and the annotated (ATG) and experimentally determined (GTG) start codons BOLD ITALIC CAPITALS. Data represents mean + standard deviation from 3 biological replicates, statistical significance by two-tailed T test (P value * < 0.1, ** < 0.01).