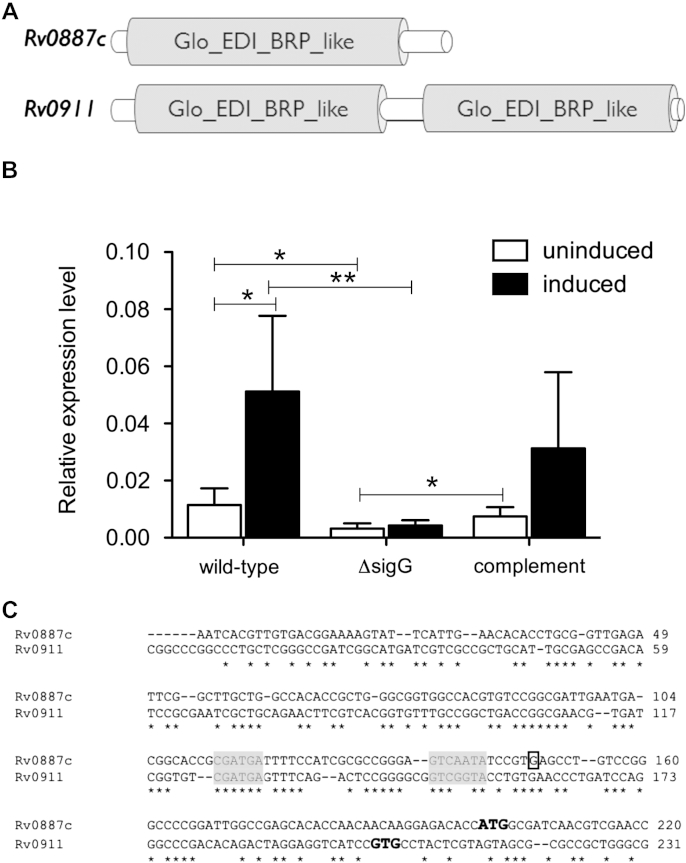
Figure 5.
SigG regulates 2 potential glyoxylases, Rv0887c and Rv0911. (A) Domain structure of Rv0887c and Rv0911 showing position of Glo_EDI_BRP_like domains. (B) Expression of Rv0887c analysed by qRT-PCR in wild-type H37Rv, ΔsigG2 mutant and ΔsigG2 complement (containing pLDL-8T) with (black bars) and without (white bars) mitomycin C induction. Data shows expression normalised to rrs, mean + standard deviation for at least 5 biological replicates, statistical significance by two-tailed T test (P value * < 0.1, ** < 0.01 and *** < 0.001). (C) 5′RACE of Rv0887c yielded a transcriptional start site, which mapped to a G residue 52 bp upstream of the annotated coding region (boxed). CLUSTAL 2.1 alignment of the upstream regions of Rv0887c and Rv0911 showed that this G residue was conserved 40 bp upstream of the annotated start codon of Rv0911 and was located in a region of strong homology between the two genes. A sigG promoter sequence CGATGA(N18)GTCNNTA was predicted (grey shading) from the alignment. Annotated start codons are shown in bold.